Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005510A_C01 KMC005510A_c01
(893 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
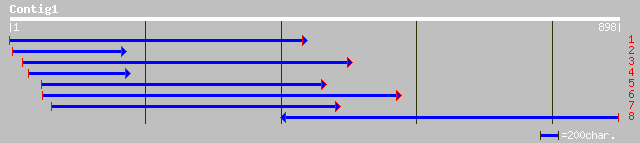
gb|AAM13095.1| unknown protein [Arabidopsis thaliana] gi|2213619... 234 1e-60
ref|NP_199428.2| 50S ribosomal protein L14; protein id: At5g4616... 234 1e-60
gb|AAL60450.1|AF402991_1 HUELLENLOS-like protein [Oryza sativa] 217 2e-55
dbj|BAB08261.1| 50S ribosomal protein L14 [Arabidopsis thaliana] 194 2e-48
gb|AAF79479.1|AC022492_23 F1L3.27 [Arabidopsis thaliana] 181 1e-44
>gb|AAM13095.1| unknown protein [Arabidopsis thaliana] gi|22136190|gb|AAM91173.1|
unknown protein [Arabidopsis thaliana]
Length = 172
Score = 234 bits (598), Expect = 1e-60
Identities = 128/172 (74%), Positives = 142/172 (82%), Gaps = 9/172 (5%)
Frame = -3
Query: 708 MAASFASRCSRVGRSLFSGLSN-SSLGSAHEMTHNNYFSQQQR-TFIQMRTVLKVVDNSG 535
MAA+FASR +R GRSL GL+N S+ S++ M + + SQQQR TFIQM TVLKVVDNSG
Sbjct: 1 MAAAFASRLTRGGRSLLGGLNNVGSMNSSNGMMNESILSQQQRRTFIQMGTVLKVVDNSG 60
Query: 534 AKKVMCIQALKGKKGARLGDTIVASVKEAHPNGKVKKGKVVYGVVVRAAMQKGRCDGGEV 355
AKKVMCIQALKGKKGARLGDTIVASVKEA PNGKVKKG VVYGVVVRAAMQ+GR DG EV
Sbjct: 61 AKKVMCIQALKGKKGARLGDTIVASVKEAMPNGKVKKGAVVYGVVVRAAMQRGRVDGSEV 120
Query: 354 RFDDNAVVLVDKQG-------QPIGTRVFGPVPHELRRKKHVQILTLPGHIA 220
RFDDNAVVLVD + QPIGTRVFGPVPHELR+KKH++IL L H+A
Sbjct: 121 RFDDNAVVLVDSKDKNTKTDRQPIGTRVFGPVPHELRKKKHLKILALAQHVA 172
>ref|NP_199428.2| 50S ribosomal protein L14; protein id: At5g46160.1, supported by
cDNA: gi_16604451, supported by cDNA: gi_18140859
[Arabidopsis thaliana] gi|16604452|gb|AAL24232.1|
AT5g46160/MCL19_22 [Arabidopsis thaliana]
gi|18140860|gb|AAL60452.1|AF402993_1 HUELLENLOS PARALOG
[Arabidopsis thaliana]
Length = 173
Score = 234 bits (597), Expect = 1e-60
Identities = 128/173 (73%), Positives = 142/173 (81%), Gaps = 10/173 (5%)
Frame = -3
Query: 708 MAASFASRCSRVGRSLFSGLSNS-SLGSAHEMTHNNYFSQQQ--RTFIQMRTVLKVVDNS 538
MAA+FASR +R GRSL GL+N S+ S++ M + + SQQQ RTFIQM TVLKVVDNS
Sbjct: 1 MAAAFASRLTRGGRSLLGGLNNGGSMNSSNGMMNESILSQQQQRRTFIQMGTVLKVVDNS 60
Query: 537 GAKKVMCIQALKGKKGARLGDTIVASVKEAHPNGKVKKGKVVYGVVVRAAMQKGRCDGGE 358
GAKKVMCIQALKGKKGARLGDTIVASVKEA PNGKVKKG VVYGVVVRAAMQ+GR DG E
Sbjct: 61 GAKKVMCIQALKGKKGARLGDTIVASVKEAMPNGKVKKGAVVYGVVVRAAMQRGRVDGSE 120
Query: 357 VRFDDNAVVLVDKQG-------QPIGTRVFGPVPHELRRKKHVQILTLPGHIA 220
VRFDDNAVVLVD + QPIGTRVFGPVPHELR+KKH++IL L H+A
Sbjct: 121 VRFDDNAVVLVDSKDKNTKTDRQPIGTRVFGPVPHELRKKKHLKILALAQHVA 173
>gb|AAL60450.1|AF402991_1 HUELLENLOS-like protein [Oryza sativa]
Length = 170
Score = 217 bits (552), Expect = 2e-55
Identities = 111/169 (65%), Positives = 133/169 (78%), Gaps = 7/169 (4%)
Frame = -3
Query: 708 MAASFASRCSRVGRSLFSGLSNSSLGSAHEM-------THNNYFSQQQRTFIQMRTVLKV 550
MAA S+CS VGR+L GL N+ G+ + +H + QQ RTFIQMRT LKV
Sbjct: 1 MAAFLRSKCSSVGRTLMGGLGNNLFGAVNSSVETVSRPSHCDPIFQQIRTFIQMRTNLKV 60
Query: 549 VDNSGAKKVMCIQALKGKKGARLGDTIVASVKEAHPNGKVKKGKVVYGVVVRAAMQKGRC 370
VDNSGAK+VMCIQ+L+GKKGARLGDTI+ SVKEA P GKVKKG VVYGVVVRAAM++GR
Sbjct: 61 VDNSGAKRVMCIQSLRGKKGARLGDTIIGSVKEAQPRGKVKKGDVVYGVVVRAAMKRGRN 120
Query: 369 DGGEVRFDDNAVVLVDKQGQPIGTRVFGPVPHELRRKKHVQILTLPGHI 223
DG E++FDDNA+VLV+ +G+ IGTRVFGPVPHELR+KKH++IL L HI
Sbjct: 121 DGSEIQFDDNAIVLVNNKGELIGTRVFGPVPHELRKKKHLKILALAEHI 169
>dbj|BAB08261.1| 50S ribosomal protein L14 [Arabidopsis thaliana]
Length = 124
Score = 194 bits (492), Expect = 2e-48
Identities = 101/124 (81%), Positives = 107/124 (85%), Gaps = 7/124 (5%)
Frame = -3
Query: 570 MRTVLKVVDNSGAKKVMCIQALKGKKGARLGDTIVASVKEAHPNGKVKKGKVVYGVVVRA 391
M TVLKVVDNSGAKKVMCIQALKGKKGARLGDTIVASVKEA PNGKVKKG VVYGVVVRA
Sbjct: 1 MGTVLKVVDNSGAKKVMCIQALKGKKGARLGDTIVASVKEAMPNGKVKKGAVVYGVVVRA 60
Query: 390 AMQKGRCDGGEVRFDDNAVVLVDKQG-------QPIGTRVFGPVPHELRRKKHVQILTLP 232
AMQ+GR DG EVRFDDNAVVLVD + QPIGTRVFGPVPHELR+KKH++IL L
Sbjct: 61 AMQRGRVDGSEVRFDDNAVVLVDSKDKNTKTDRQPIGTRVFGPVPHELRKKKHLKILALA 120
Query: 231 GHIA 220
H+A
Sbjct: 121 QHVA 124
>gb|AAF79479.1|AC022492_23 F1L3.27 [Arabidopsis thaliana]
Length = 201
Score = 181 bits (459), Expect = 1e-44
Identities = 109/197 (55%), Positives = 134/197 (67%), Gaps = 34/197 (17%)
Frame = -3
Query: 711 KMAASFASRCSRVGRSLFSGLSNSSLG----SAHEMTHNNYFSQQQ-RTFIQMRTVLKVV 547
+MA + AS+ S+ GRSL GL N+ G S++ M + + SQQQ RTFIQM T+LK V
Sbjct: 5 EMATALASKLSK-GRSLLGGLCNAFSGLMNSSSNGMMNGSILSQQQHRTFIQMGTILKCV 63
Query: 546 DNSGAKKVMCIQALKGKKGARLGDTIVASVKEAH------------PNGKVKKGKVVYGV 403
DNS AK+VMCIQ+L+GKKGARLGD IV SVKEA+ P GKVKKG VVYGV
Sbjct: 64 DNSCAKEVMCIQSLRGKKGARLGDIIVGSVKEANPIVQKKVKKDAIPKGKVKKGMVVYGV 123
Query: 402 VVRAAMQKGRCDGGEVRFDDNAVVLV---DKQG--------------QPIGTRVFGPVPH 274
VVRAAM KGR DG +V+FDDNA+V+V +K+G QP GTRVFGPVPH
Sbjct: 124 VVRAAMPKGRADGSQVKFDDNAIVVVGIKEKKGQNNSHGSKRKMEYNQPTGTRVFGPVPH 183
Query: 273 ELRRKKHVQILTLPGHI 223
E+R +K ++IL+L HI
Sbjct: 184 EMRLRKQLKILSLAQHI 200
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 806,513,541
Number of Sequences: 1393205
Number of extensions: 18633222
Number of successful extensions: 49185
Number of sequences better than 10.0: 282
Number of HSP's better than 10.0 without gapping: 46294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48893
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 48775691475
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)