Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005492A_C01 KMC005492A_c01
(578 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
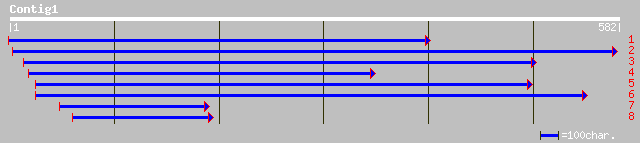
ref|NP_566899.1| putative protein; protein id: At3g48200.1, supp... 96 2e-19
gb|AAK64034.1| unknown protein [Arabidopsis thaliana] 96 2e-19
pir||T13009 hypothetical protein T24C20.80 - Arabidopsis thalian... 94 9e-19
ref|NP_501354.1| Golgi protein, possibly N-myristoylated (53.7 k... 39 0.061
ref|NP_628969.1| putative integral membrane protein [Streptomyce... 36 0.40
>ref|NP_566899.1| putative protein; protein id: At3g48200.1, supported by cDNA:
gi_14532611 [Arabidopsis thaliana]
Length = 1088
Score = 96.3 bits (238), Expect = 2e-19
Identities = 67/141 (47%), Positives = 85/141 (59%), Gaps = 13/141 (9%)
Frame = -3
Query: 576 KSFLKVMKIASIGILVYFFPQNCTKH------LEKKLPQNGHGNGDTGLERYR-SSGSRS 418
+SFL +K+ IG+ PQ K+ LE + NG G T RYR SSGSRS
Sbjct: 953 RSFLSGLKMFIIGLAALILPQKMIKNKIPVAQLEARSSSNG---GTTPEFRYRNSSGSRS 1009
Query: 417 SGGTPDKPWLKRLRELGKLSFGRERSGA--INDPSTSSTTRWSGFWGNKRSGS----SSS 256
SG DKPWLK++RE+ K SF R+RS + +DPS S + S WG K SGS SS+
Sbjct: 1010 SGSL-DKPWLKQIREMAKSSFTRDRSNSKVPSDPSCSKSGWSSSIWGTKTSGSSSKESSA 1068
Query: 255 DFKSKPSSTLHQDLEAIFASK 193
D+KS+P L++DLEAIFASK
Sbjct: 1069 DYKSRPKG-LYKDLEAIFASK 1088
>gb|AAK64034.1| unknown protein [Arabidopsis thaliana]
Length = 1088
Score = 96.3 bits (238), Expect = 2e-19
Identities = 67/141 (47%), Positives = 85/141 (59%), Gaps = 13/141 (9%)
Frame = -3
Query: 576 KSFLKVMKIASIGILVYFFPQNCTKH------LEKKLPQNGHGNGDTGLERYR-SSGSRS 418
+SFL +K+ IG+ PQ K+ LE + NG G T RYR SSGSRS
Sbjct: 953 RSFLSGLKMFIIGLAALILPQKMIKNKIPVAQLEARSSSNG---GTTPEFRYRNSSGSRS 1009
Query: 417 SGGTPDKPWLKRLRELGKLSFGRERSGA--INDPSTSSTTRWSGFWGNKRSGS----SSS 256
SG DKPWLK++RE+ K SF R+RS + +DPS S + S WG K SGS SS+
Sbjct: 1010 SGSL-DKPWLKQIREMAKSSFTRDRSNSKVPSDPSCSKSGWSSSIWGTKTSGSSSKESSA 1068
Query: 255 DFKSKPSSTLHQDLEAIFASK 193
D+KS+P L++DLEAIFASK
Sbjct: 1069 DYKSRPKG-LYKDLEAIFASK 1088
>pir||T13009 hypothetical protein T24C20.80 - Arabidopsis thaliana
gi|5541726|emb|CAB51067.1| putative protein [Arabidopsis
thaliana]
Length = 1998
Score = 94.4 bits (233), Expect = 9e-19
Identities = 66/140 (47%), Positives = 84/140 (59%), Gaps = 13/140 (9%)
Frame = -3
Query: 576 KSFLKVMKIASIGILVYFFPQNCTKH------LEKKLPQNGHGNGDTGLERYR-SSGSRS 418
+SFL +K+ IG+ PQ K+ LE + NG G T RYR SSGSRS
Sbjct: 870 RSFLSGLKMFIIGLAALILPQKMIKNKIPVAQLEARSSSNG---GTTPEFRYRNSSGSRS 926
Query: 417 SGGTPDKPWLKRLRELGKLSFGRERSGA--INDPSTSSTTRWSGFWGNKRSGS----SSS 256
SG DKPWLK++RE+ K SF R+RS + +DPS S + S WG K SGS SS+
Sbjct: 927 SGSL-DKPWLKQIREMAKSSFTRDRSNSKVPSDPSCSKSGWSSSIWGTKTSGSSSKESSA 985
Query: 255 DFKSKPSSTLHQDLEAIFAS 196
D+KS+P L++DLEAIFAS
Sbjct: 986 DYKSRPKG-LYKDLEAIFAS 1004
>ref|NP_501354.1| Golgi protein, possibly N-myristoylated (53.7 kD) [Caenorhabditis
elegans] gi|7331876|gb|AAF60564.1| Hypothetical protein
Y42H9AR.1 [Caenorhabditis elegans]
Length = 506
Score = 38.5 bits (88), Expect = 0.061
Identities = 26/76 (34%), Positives = 38/76 (49%), Gaps = 5/76 (6%)
Frame = +1
Query: 268 PTPFVSPETTPSSGATGRRI-IYSSTSFPTKAKLSKFPQSFQPWFIRCTT*APTP*A--- 435
PT + P + +S ++G + YS S+PT+ + S P + P F T APTP
Sbjct: 392 PTSYYPPAASTASNSSGYQAPYYSPPSYPTQQQSSYVPSAVPPMF--SATSAPTPPTSYV 449
Query: 436 -PIPFQTSVSIPMPIL 480
PIP ++ PMP L
Sbjct: 450 NPIPPPAPLNFPMPSL 465
>ref|NP_628969.1| putative integral membrane protein [Streptomyces coelicolor A3(2)]
gi|8218213|emb|CAB92675.1| putative integral membrane
protein [Streptomyces coelicolor A3(2)]
Length = 265
Score = 35.8 bits (81), Expect = 0.40
Identities = 21/81 (25%), Positives = 31/81 (37%)
Frame = -3
Query: 471 HGNGDTGLERYRSSGSRSSGGTPDKPWLKRLRELGKLSFGRERSGAINDPSTSSTTRWSG 292
+G G G + + GTPD PW R S + GA + W G
Sbjct: 42 NGAGQDGSSDQDNPFAPPPEGTPDSPWQPR-----HPSGDGDSQGASGSQGEGGRSPWGG 96
Query: 291 FWGNKRSGSSSSDFKSKPSST 229
W +++ G S F +PS +
Sbjct: 97 RWSDRQPGRSPGGFGERPSGS 117
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 537,130,795
Number of Sequences: 1393205
Number of extensions: 12384815
Number of successful extensions: 34930
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 33085
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34805
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21426319650
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)