Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005491A_C01 KMC005491A_c01
(689 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
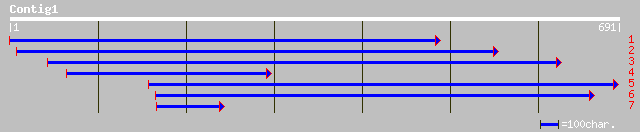
ref|NP_194745.1| putative villin; protein id: At4g30160.1 [Arabi... 100 3e-20
sp|O81643|VIL1_ARATH Villin 1 gi|11358918|pir||T50671 villin 1 [... 99 4e-20
ref|NP_029567.1| putative villin; protein id: At2g29890.1, suppo... 99 6e-20
pir||H84701 probable villin [imported] - Arabidopsis thaliana 99 6e-20
dbj|BAC42808.1| putative villin 1 VLN1 [Arabidopsis thaliana] 99 6e-20
>ref|NP_194745.1| putative villin; protein id: At4g30160.1 [Arabidopsis thaliana]
gi|25091517|sp|O65570|VIL4_ARATH Villin 4
gi|7488222|pir||T14076 probable villin [imported] -
Arabidopsis thaliana gi|3093294|emb|CAA73320.1| putative
villin [Arabidopsis thaliana] gi|5730126|emb|CAB52460.1|
putative villin [Arabidopsis thaliana]
gi|7269916|emb|CAB81009.1| putative villin [Arabidopsis
thaliana] gi|26449688|dbj|BAC41968.1| putative villin
[Arabidopsis thaliana] gi|29029072|gb|AAO64915.1|
At4g30160 [Arabidopsis thaliana]
Length = 974
Score = 99.8 bits (247), Expect = 3e-20
Identities = 66/179 (36%), Positives = 90/179 (49%), Gaps = 53/179 (29%)
Frame = -2
Query: 679 FRQSGDRLLSSPSPVVKKLFDGS---------PANSSA---------------EQTLPQA 572
F R LS+P PVV+KL+ S PA S+ E ++P+
Sbjct: 797 FESQNARNLSTPPPVVRKLYPRSVTPDSSKFAPAPKSSAIASRSALFEKIPPQEPSIPKP 856
Query: 571 --------DSPATELSSSNERESFTQKDRNV---------------------DGENLLVY 479
+SPA E S+S E+E + D+ D E+L +
Sbjct: 857 VKASPKTPESPAPE-SNSKEQEEKKENDKEEGSMSSRIESLTIQEDAKEGVEDEEDLPAH 915
Query: 478 PYEHLRVVSANPVTGIDITRRETYLSNEEFHEKFGMPKSAFYKLPRWKQNKLKMSLDLY 302
PY+ L+ S +PV+ ID+TRRE YLS+EEF EKFGM K AFYKLP+WKQNK KM++ L+
Sbjct: 916 PYDRLKTTSTDPVSDIDVTRREAYLSSEEFKEKFGMTKEAFYKLPKWKQNKFKMAVQLF 974
>sp|O81643|VIL1_ARATH Villin 1 gi|11358918|pir||T50671 villin 1 [imported] - Arabidopsis
thaliana gi|3415113|gb|AAC31605.1| villin 1 [Arabidopsis
thaliana]
Length = 910
Score = 99.4 bits (246), Expect = 4e-20
Identities = 54/117 (46%), Positives = 69/117 (58%)
Frame = -2
Query: 652 SSPSPVVKKLFDGSPANSSAEQTLPQADSPATELSSSNERESFTQKDRNVDGENLLVYPY 473
S+ +PVVKKLF S + Q S +++S R +++ L Y Y
Sbjct: 797 SNSTPVVKKLFSESLVVDPNDGVARQESSSKSDISKQKPRVGINSDLSSLES---LAYSY 853
Query: 472 EHLRVVSANPVTGIDITRRETYLSNEEFHEKFGMPKSAFYKLPRWKQNKLKMSLDLY 302
E LRV S PVT ID TRRE YL+ +EF E+FGM KS FY LP+WKQNKLK+SL L+
Sbjct: 854 EQLRVDSQKPVTDIDATRREAYLTEKEFEERFGMAKSEFYALPKWKQNKLKISLHLF 910
>ref|NP_029567.1| putative villin; protein id: At2g29890.1, supported by cDNA:
gi_3415112 [Arabidopsis thaliana]
gi|20198093|gb|AAD23629.2| putative villin [Arabidopsis
thaliana]
Length = 909
Score = 99.0 bits (245), Expect = 6e-20
Identities = 54/117 (46%), Positives = 69/117 (58%)
Frame = -2
Query: 652 SSPSPVVKKLFDGSPANSSAEQTLPQADSPATELSSSNERESFTQKDRNVDGENLLVYPY 473
S+ +PVVKKLF S + Q S +++S R +++ L Y Y
Sbjct: 796 SNSTPVVKKLFSESLLVDPNDGVARQESSSKSDISKQKPRVGINSDLSSLES---LAYSY 852
Query: 472 EHLRVVSANPVTGIDITRRETYLSNEEFHEKFGMPKSAFYKLPRWKQNKLKMSLDLY 302
E LRV S PVT ID TRRE YL+ +EF E+FGM KS FY LP+WKQNKLK+SL L+
Sbjct: 853 EQLRVDSQKPVTDIDATRREAYLTEKEFEERFGMAKSEFYALPKWKQNKLKISLHLF 909
>pir||H84701 probable villin [imported] - Arabidopsis thaliana
Length = 898
Score = 99.0 bits (245), Expect = 6e-20
Identities = 54/117 (46%), Positives = 69/117 (58%)
Frame = -2
Query: 652 SSPSPVVKKLFDGSPANSSAEQTLPQADSPATELSSSNERESFTQKDRNVDGENLLVYPY 473
S+ +PVVKKLF S + Q S +++S R +++ L Y Y
Sbjct: 785 SNSTPVVKKLFSESLLVDPNDGVARQESSSKSDISKQKPRVGINSDLSSLES---LAYSY 841
Query: 472 EHLRVVSANPVTGIDITRRETYLSNEEFHEKFGMPKSAFYKLPRWKQNKLKMSLDLY 302
E LRV S PVT ID TRRE YL+ +EF E+FGM KS FY LP+WKQNKLK+SL L+
Sbjct: 842 EQLRVDSQKPVTDIDATRREAYLTEKEFEERFGMAKSEFYALPKWKQNKLKISLHLF 898
>dbj|BAC42808.1| putative villin 1 VLN1 [Arabidopsis thaliana]
Length = 718
Score = 99.0 bits (245), Expect = 6e-20
Identities = 54/117 (46%), Positives = 69/117 (58%)
Frame = -2
Query: 652 SSPSPVVKKLFDGSPANSSAEQTLPQADSPATELSSSNERESFTQKDRNVDGENLLVYPY 473
S+ +PVVKKLF S + Q S +++S R +++ L Y Y
Sbjct: 605 SNSTPVVKKLFSESLLVDPNDGVARQESSSKSDISKQKPRVGINSDLSSLES---LAYSY 661
Query: 472 EHLRVVSANPVTGIDITRRETYLSNEEFHEKFGMPKSAFYKLPRWKQNKLKMSLDLY 302
E LRV S PVT ID TRRE YL+ +EF E+FGM KS FY LP+WKQNKLK+SL L+
Sbjct: 662 EQLRVDSQKPVTDIDATRREAYLTEKEFEERFGMAKSEFYALPKWKQNKLKISLHLF 718
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 568,993,577
Number of Sequences: 1393205
Number of extensions: 11843260
Number of successful extensions: 33198
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 32267
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33177
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31118763720
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)