Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005481A_C01 KMC005481A_c01
(674 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
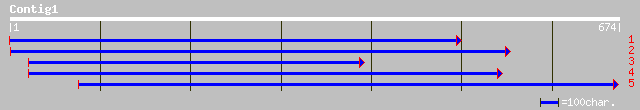
ref|NP_190834.1| RRM-containing RNA-binding protein, putative; p... 112 6e-24
pir||T01563 hypothetical protein A_TM018A10.14 - Arabidopsis tha... 102 6e-21
ref|NP_567192.1| RRM-containing protein; protein id: At4g00830.1... 102 6e-21
gb|AAL69426.1|AC098565_8 Putative RNA-binding protein [Oryza sat... 102 6e-21
gb|AAO72701.1| putative RNA-binding protein [Oryza sativa (japon... 85 8e-16
>ref|NP_190834.1| RRM-containing RNA-binding protein, putative; protein id:
At3g52660.1 [Arabidopsis thaliana]
gi|11358672|pir||T49019 probable RNA binding protein -
Arabidopsis thaliana gi|7669940|emb|CAB89227.1| putative
RNA binding protein [Arabidopsis thaliana]
Length = 471
Score = 112 bits (279), Expect = 6e-24
Identities = 66/135 (48%), Positives = 80/135 (58%), Gaps = 6/135 (4%)
Frame = -3
Query: 672 DQKPG---VLHTQKQGLLPRYPPSVGYGLLGAPYGALGAGYGAPGLAQPFMYGAGQTPPG 502
DQK V + QK L P YPP + YG+ +P+GALG G+GA +QP M+ G G
Sbjct: 349 DQKTNTNTVQNVQKSQLQPNYPPLLSYGMAPSPFGALG-GFGASAYSQPLMHAGGHAAGG 407
Query: 501 MSMMPMLLADGRIGYVLQQPGL--QPQVPPPSHQRGGRSGGGGDSGGSSNRNSGSSSKGR 328
MSMMP++L DGRIGYVLQQPGL PQ PPP H R G G S SSS +
Sbjct: 408 MSMMPIMLPDGRIGYVLQQPGLAAMPQ-PPPRHSPPYRGGSG----------SSSSSGSK 456
Query: 327 HNNDGGQGR-RYRPY 286
++D G+GR RY PY
Sbjct: 457 RSSDNGRGRSRYNPY 471
>pir||T01563 hypothetical protein A_TM018A10.14 - Arabidopsis thaliana
gi|2252863|gb|AAB62861.1| similar to nucleolin protein
[Arabidopsis thaliana] gi|7267422|emb|CAB80892.1|
putative protein [Arabidopsis thaliana]
Length = 521
Score = 102 bits (253), Expect = 6e-21
Identities = 58/116 (50%), Positives = 67/116 (57%), Gaps = 2/116 (1%)
Frame = -3
Query: 627 PRYPPSVGYGLLGAPYGALGAGYGAPG-LAQPFMYGAGQTPPGMSMMPMLLADGRIGYVL 451
P P+ G G APYGA+GAG G G +QP +YG G P GM M+PMLL DGR+GYVL
Sbjct: 415 PFVHPTFG-GFAAAPYGAMGAGLGIAGSFSQPMIYGRGAMPTGMQMVPMLLPDGRVGYVL 473
Query: 450 QQPGLQPQVPPPSHQRGGRSGGGGDSGGSSNRNSGSSSKGRHNNDGGQ-GRRYRPY 286
QQPG+ PP R G SGGS NS H +DG + GRRYRPY
Sbjct: 474 QQPGMPMAAAPPQRPR-RNDRNNGSSGGSGRDNS-------HEHDGNRGGRRYRPY 521
>ref|NP_567192.1| RRM-containing protein; protein id: At4g00830.1, supported by cDNA:
gi_13605915 [Arabidopsis thaliana]
gi|13605916|gb|AAK32943.1|AF367357_1
AT4g00830/A_TM018A10_14 [Arabidopsis thaliana]
gi|21360555|gb|AAM47474.1| AT4g00830/A_TM018A10_14
[Arabidopsis thaliana]
Length = 495
Score = 102 bits (253), Expect = 6e-21
Identities = 58/116 (50%), Positives = 67/116 (57%), Gaps = 2/116 (1%)
Frame = -3
Query: 627 PRYPPSVGYGLLGAPYGALGAGYGAPG-LAQPFMYGAGQTPPGMSMMPMLLADGRIGYVL 451
P P+ G G APYGA+GAG G G +QP +YG G P GM M+PMLL DGR+GYVL
Sbjct: 389 PFVHPTFG-GFAAAPYGAMGAGLGIAGSFSQPMIYGRGAMPTGMQMVPMLLPDGRVGYVL 447
Query: 450 QQPGLQPQVPPPSHQRGGRSGGGGDSGGSSNRNSGSSSKGRHNNDGGQ-GRRYRPY 286
QQPG+ PP R G SGGS NS H +DG + GRRYRPY
Sbjct: 448 QQPGMPMAAAPPQRPR-RNDRNNGSSGGSGRDNS-------HEHDGNRGGRRYRPY 495
>gb|AAL69426.1|AC098565_8 Putative RNA-binding protein [Oryza sativa]
Length = 472
Score = 102 bits (253), Expect = 6e-21
Identities = 59/129 (45%), Positives = 69/129 (52%)
Frame = -3
Query: 672 DQKPGVLHTQKQGLLPRYPPSVGYGLLGAPYGALGAGYGAPGLAQPFMYGAGQTPPGMSM 493
D+KP H+ K G P +P G +G PYGA G G PG QP +YG G P GM M
Sbjct: 367 DKKPD--HSFKPGGAPNFPLPPYGGYMGDPYGAYGGG--GPGFNQPMIYGRGPAPAGMRM 422
Query: 492 MPMLLADGRIGYVLQQPGLQPQVPPPSHQRGGRSGGGGDSGGSSNRNSGSSSKGRHNNDG 313
+PM+L DGR+GYVLQQPG P PPP +RG R GG G +G
Sbjct: 423 VPMVLPDGRLGYVLQQPGGIP--PPPPPRRGDRRDGGSRGG-----------------EG 463
Query: 312 GQGRRYRPY 286
GRRYRPY
Sbjct: 464 SHGRRYRPY 472
>gb|AAO72701.1| putative RNA-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 476
Score = 85.1 bits (209), Expect = 8e-16
Identities = 49/91 (53%), Positives = 60/91 (65%), Gaps = 1/91 (1%)
Frame = -3
Query: 633 LLPRYPPSVGYGLLGAPYGALGAGYGAPGLAQPFMYGAGQTPPGMSMMPMLLADGRIGYV 454
LLP YPP +GYG++ P GA GA + AQP +Y A + PPG +M+PM+L DGR+ YV
Sbjct: 391 LLPSYPP-LGYGIMSVP-GAYGAAPAST--AQPMLY-APRAPPGAAMVPMMLPDGRLVYV 445
Query: 453 LQQPGLQ-PQVPPPSHQRGGRSGGGGDSGGS 364
+QQPG Q P PP Q G RSG GG GGS
Sbjct: 446 VQQPGGQLPLASPPPQQAGHRSGSGGRHGGS 476
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 670,608,684
Number of Sequences: 1393205
Number of extensions: 18425147
Number of successful extensions: 165401
Number of sequences better than 10.0: 1244
Number of HSP's better than 10.0 without gapping: 76905
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 134043
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29704274460
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)