Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005458A_C01 KMC005458A_c01
(600 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
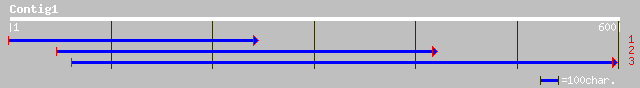
ref|NP_568832.1| expressed protein; protein id: At5g55940.1, sup... 105 6e-22
gb|AAM64789.1| unknown [Arabidopsis thaliana] 105 6e-22
gb|AAN01224.1| chlorophyll antenna size regulatory protein [Chla... 46 3e-04
ref|XP_226537.1| similar to Neighbor of COX4 [Rattus norvegicus] 44 0.002
ref|NP_035056.1| neighbor of Cox4 [Mus musculus] gi|18202123|sp|... 44 0.002
>ref|NP_568832.1| expressed protein; protein id: At5g55940.1, supported by cDNA:
33108., supported by cDNA: gi_18086570 [Arabidopsis
thaliana] gi|18202899|sp|Q9FG71|U172_ARATH Hypothetical
protein At5g55940 gi|9758214|dbj|BAB08659.1|
gene_id:MYN21.5~unknown protein [Arabidopsis thaliana]
gi|18086571|gb|AAL57709.1| AT5g55940/MYN21_5
[Arabidopsis thaliana] gi|21689597|gb|AAM67420.1|
AT5g55940/MYN21_5 [Arabidopsis thaliana]
Length = 208
Score = 105 bits (261), Expect = 6e-22
Identities = 45/65 (69%), Positives = 55/65 (84%)
Frame = -3
Query: 598 LYLRDASKNWKLAQPDGNSRFSLKEPSANLVLSDYISSEKWNNIVDFDDHLDDISKDWLN 419
L ++DASKNW++ DG S+ LKEPSAN+VLSDYISSEKW ++ D DDHLDD++KDWLN
Sbjct: 144 LCVKDASKNWRVVGADGGSKLLLKEPSANVVLSDYISSEKWKDVTDVDDHLDDVTKDWLN 203
Query: 418 PGLFN 404
PGLFN
Sbjct: 204 PGLFN 208
>gb|AAM64789.1| unknown [Arabidopsis thaliana]
Length = 206
Score = 105 bits (261), Expect = 6e-22
Identities = 45/65 (69%), Positives = 55/65 (84%)
Frame = -3
Query: 598 LYLRDASKNWKLAQPDGNSRFSLKEPSANLVLSDYISSEKWNNIVDFDDHLDDISKDWLN 419
L ++DASKNW++ DG S+ LKEPSAN+VLSDYISSEKW ++ D DDHLDD++KDWLN
Sbjct: 142 LCVKDASKNWRVVGADGGSKLLLKEPSANVVLSDYISSEKWKDVTDVDDHLDDVTKDWLN 201
Query: 418 PGLFN 404
PGLFN
Sbjct: 202 PGLFN 206
>gb|AAN01224.1| chlorophyll antenna size regulatory protein [Chlamydomonas
reinhardtii] gi|22536150|gb|AAN01225.1| chlorophyll
antenna size regulatory protein [Chlamydomonas
reinhardtii]
Length = 213
Score = 46.2 bits (108), Expect = 3e-04
Identities = 24/65 (36%), Positives = 33/65 (49%), Gaps = 3/65 (4%)
Frame = -3
Query: 598 LYLRDASKNWKLAQPDGNSRFSLKEPSANLVLSDYI---SSEKWNNIVDFDDHLDDISKD 428
L+ +D SK WK A DG +LK + ++ K + DF++HLDD KD
Sbjct: 136 LFSKDGSKGWKRASADGGE-LALKNADWKKLREEFFVMFKQLKHRTLHDFEEHLDDAGKD 194
Query: 427 WLNPG 413
WLN G
Sbjct: 195 WLNKG 199
>ref|XP_226537.1| similar to Neighbor of COX4 [Rattus norvegicus]
Length = 262
Score = 43.5 bits (101), Expect = 0.002
Identities = 18/45 (40%), Positives = 25/45 (55%)
Frame = -3
Query: 523 PSANLVLSDYISSEKWNNIVDFDDHLDDISKDWLNPGLFN*CQHL 389
P A + + + S + +VDFD+HLDDI DW NP + HL
Sbjct: 217 PEAQRISASLLDSRSYETLVDFDNHLDDIRSDWTNPEINKAVLHL 261
>ref|NP_035056.1| neighbor of Cox4 [Mus musculus] gi|18202123|sp|O70378|NOC4_MOUSE
Neighbor of COX4 gi|3037045|gb|AAC12933.1| hypothetical
protein COX4AL [Mus musculus] gi|14318610|gb|AAH09103.1|
neighbor of Cox4 [Mus musculus]
Length = 207
Score = 43.5 bits (101), Expect = 0.002
Identities = 18/45 (40%), Positives = 25/45 (55%)
Frame = -3
Query: 523 PSANLVLSDYISSEKWNNIVDFDDHLDDISKDWLNPGLFN*CQHL 389
P A + + + S + +VDFD+HLDDI DW NP + HL
Sbjct: 162 PEAQRISASLLDSRSYETLVDFDNHLDDIRSDWTNPEINKAVLHL 206
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 503,844,570
Number of Sequences: 1393205
Number of extensions: 10844654
Number of successful extensions: 28428
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 27771
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28420
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)