Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005450A_C01 KMC005450A_c01
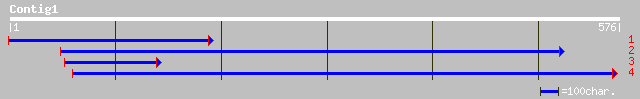
(576 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172129.1| trehalose-6-phosphate synthase, putative; prote... 193 1e-48
gb|AAL91978.1|AF483209_1 putative trehalose synthase [Solanum tu... 192 3e-48
dbj|BAB56048.1| putative trehalose-6-phosphate synthase homolog ... 182 2e-45
dbj|BAB21172.1| putative trehalose-6-phosphate synthase [Oryza s... 178 4e-44
ref|NP_567538.1| trehalose-6-phosphate synthase like protein; pr... 164 7e-40
>ref|NP_172129.1| trehalose-6-phosphate synthase, putative; protein id: At1g06410.1
[Arabidopsis thaliana] gi|25347117|pir||A86200
hypothetical protein [imported] - Arabidopsis thaliana
gi|8927678|gb|AAF82169.1|AC068143_11 Contains similarity
to a trehalose-6-phosphate synthase mRNA from Arabidopsis
thaliana gb|Y08568 and contains a trehalose-6-phosphate
synthase PF|00982 domain. ESTs gb|T76758, gb|T21695,
gb|R30506, gb|T42298, gb|T42288 come from this gene
Length = 851
Score = 193 bits (491), Expect = 1e-48
Identities = 97/125 (77%), Positives = 109/125 (86%)
Frame = -1
Query: 576 AKEMLDHLESVLANEPVAVKSGQFIVEVKPQDVSKGLVAEKIFSSMAGNGKPADFVLCVG 397
AKEML+HLESVLANEPVAVKSG +IVEVKPQ VSKG V+EKIFSSMAG GKP DFVLC+G
Sbjct: 718 AKEMLEHLESVLANEPVAVKSGHYIVEVKPQGVSKGSVSEKIFSSMAGKGKPVDFVLCIG 777
Query: 396 DDRSDEDMFEIVRSAISRNILSPNASVFACTVGQKPSKAKYYLDDTFEVINMLESLAEES 217
DDRSDEDMFE + +A+S+ +L NA VFACTVGQKPSKAKYYLDDT EV MLESLAE S
Sbjct: 778 DDRSDEDMFEAIGNAMSKRLLCDNALVFACTVGQKPSKAKYYLDDTTEVTCMLESLAEAS 837
Query: 216 DSSPY 202
++S +
Sbjct: 838 EASNF 842
>gb|AAL91978.1|AF483209_1 putative trehalose synthase [Solanum tuberosum]
Length = 857
Score = 192 bits (488), Expect = 3e-48
Identities = 96/122 (78%), Positives = 110/122 (89%)
Frame = -1
Query: 576 AKEMLDHLESVLANEPVAVKSGQFIVEVKPQDVSKGLVAEKIFSSMAGNGKPADFVLCVG 397
AKEMLDHLESVLANEPVAVKSGQ IVEVKPQ V+KGLVAEK+F+S+A GK ADFVLC+G
Sbjct: 724 AKEMLDHLESVLANEPVAVKSGQHIVEVKPQGVTKGLVAEKVFTSLAVKGKLADFVLCIG 783
Query: 396 DDRSDEDMFEIVRSAISRNILSPNASVFACTVGQKPSKAKYYLDDTFEVINMLESLAEES 217
DDRSDEDMFEI+ A+SRNI+S +A VFACTVGQKPSKAKYYLDDT EV+ ML+SLA+ +
Sbjct: 784 DDRSDEDMFEIIGDALSRNIISYDAKVFACTVGQKPSKAKYYLDDTSEVVLMLDSLADAT 843
Query: 216 DS 211
D+
Sbjct: 844 DT 845
>dbj|BAB56048.1| putative trehalose-6-phosphate synthase homolog [Oryza sativa
(japonica cultivar-group)]
Length = 912
Score = 182 bits (463), Expect = 2e-45
Identities = 89/131 (67%), Positives = 107/131 (80%)
Frame = -1
Query: 576 AKEMLDHLESVLANEPVAVKSGQFIVEVKPQDVSKGLVAEKIFSSMAGNGKPADFVLCVG 397
AKE+LDHLESVLANEPV+VKSGQFIVEVKPQ VSKG+VAEKI SM GK ADFVLC+G
Sbjct: 756 AKELLDHLESVLANEPVSVKSGQFIVEVKPQGVSKGVVAEKILVSMKERGKQADFVLCIG 815
Query: 396 DDRSDEDMFEIVRSAISRNILSPNASVFACTVGQKPSKAKYYLDDTFEVINMLESLAEES 217
DDRSDEDMFE + I + +++ N S+FACTVGQKPSKAK+YLDDTFEV+ ML +LA+ +
Sbjct: 816 DDRSDEDMFENIADTIKKGMVATNTSLFACTVGQKPSKAKFYLDDTFEVVTMLSALADAT 875
Query: 216 DSSPYIEETGD 184
+ P + T +
Sbjct: 876 EPEPETDLTDE 886
>dbj|BAB21172.1| putative trehalose-6-phosphate synthase [Oryza sativa (japonica
cultivar-group)] gi|15289825|dbj|BAB63523.1| putative
trehalose-6-phosphate synthase [Oryza sativa (japonica
cultivar-group)]
Length = 878
Score = 178 bits (452), Expect = 4e-44
Identities = 92/131 (70%), Positives = 106/131 (80%), Gaps = 3/131 (2%)
Frame = -1
Query: 576 AKEMLDHLESVLANEPVAVKSGQFIVEVKPQDVSKGLVAEKIFSSMAGNGKPADFVLCVG 397
AKEMLDHLESVLANEPV VKSGQ IVEVKPQ VSKG VAEKI S++ N + ADFVLC+G
Sbjct: 740 AKEMLDHLESVLANEPVCVKSGQQIVEVKPQGVSKGFVAEKILSTLTENKRQADFVLCIG 799
Query: 396 DDRSDEDMFEIVRSAISRNILSPNASVFACTVGQKPSKAKYYLDDTFEVINMLESLA--- 226
DDRSDEDMFE + + R+I+ P S++ACTVGQKPSKAKYYLDDT +V+NMLE+LA
Sbjct: 800 DDRSDEDMFEGIADIMRRSIVDPQTSLYACTVGQKPSKAKYYLDDTNDVLNMLEALADAS 859
Query: 225 EESDSSPYIEE 193
EE+DS EE
Sbjct: 860 EETDSQEDAEE 870
>ref|NP_567538.1| trehalose-6-phosphate synthase like protein; protein id: At4g17770.1,
supported by cDNA: 95947. [Arabidopsis thaliana]
gi|26452424|dbj|BAC43297.1| putative
trehalose-6-phosphate synthase [Arabidopsis thaliana]
gi|29029046|gb|AAO64902.1| At4g17770 [Arabidopsis
thaliana]
Length = 862
Score = 164 bits (415), Expect = 7e-40
Identities = 79/117 (67%), Positives = 97/117 (82%)
Frame = -1
Query: 576 AKEMLDHLESVLANEPVAVKSGQFIVEVKPQDVSKGLVAEKIFSSMAGNGKPADFVLCVG 397
AKE+++HLESVL N+PV+VK+GQ +VEVKPQ V+KGLVAE++ ++M GK DF+LCVG
Sbjct: 724 AKELMEHLESVLTNDPVSVKTGQQLVEVKPQGVNKGLVAERLLTTMQEKGKLLDFILCVG 783
Query: 396 DDRSDEDMFEIVRSAISRNILSPNASVFACTVGQKPSKAKYYLDDTFEVINMLESLA 226
DDRSDEDMFE++ SA LSP A +FACTVGQKPSKAKYYLDDT E+I ML+ LA
Sbjct: 784 DDRSDEDMFEVIMSAKDGPALSPVAEIFACTVGQKPSKAKYYLDDTAEIIRMLDGLA 840
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 477,190,660
Number of Sequences: 1393205
Number of extensions: 10069645
Number of successful extensions: 27943
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 26955
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27888
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)