Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
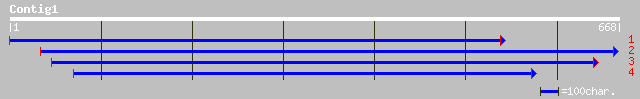
Query= KMC005438A_C01 KMC005438A_c01
(630 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC41892.1| unknown protein [Arabidopsis thaliana] 264 8e-70
ref|NP_198037.1| putative protein; protein id: At5g26850.1 [Arab... 264 8e-70
gb|AAK56279.1|AF367291_1 At1g05960/T21E18_20 [Arabidopsis thalia... 191 9e-48
pir||F86194 hypothetical protein [imported] - Arabidopsis thalia... 191 9e-48
ref|NP_563755.1| expressed protein; protein id: At1g05960.1, sup... 191 9e-48
>dbj|BAC41892.1| unknown protein [Arabidopsis thaliana]
Length = 970
Score = 264 bits (674), Expect = 8e-70
Identities = 124/178 (69%), Positives = 147/178 (81%)
Frame = +1
Query: 97 MGIISRKIFPACGNMCVCCPALRSSSRQPVKRYKKLLADIFPKSPDELPSERKIVKLCEY 276
MG ISR +FPAC +MC+CCPALRS SRQPVKRYKKLL +IFPKSPD P+ERKIVKLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 277 AAKNPFRIPKIAKYLEERCYKELRSGHIKLVTIITESFNKLLSICKVQIAYFALDVLNVI 456
AAKNP RIPKIAK+LEERCYK+LRS +K + I+TE++NK+L CK Q+AYFA +LNV+
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 457 TELLGYSKDETIQTLGCQSLTRFIFCQVDTTYTHNIEKLVRKVCILSREHGETHEKRC 630
TELL SK +T LGCQ+LTRFI+ QVD TYTH+IEK KVC L+RE GE H+K+C
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQC 178
>ref|NP_198037.1| putative protein; protein id: At5g26850.1 [Arabidopsis thaliana]
gi|7485250|pir||T01764 hypothetical protein
A_IG002P16.24 - Arabidopsis thaliana
gi|2191175|gb|AAB61061.1| Hypothetical protein F2P16.24
[Arabidopsis thaliana]
Length = 907
Score = 264 bits (674), Expect = 8e-70
Identities = 124/178 (69%), Positives = 147/178 (81%)
Frame = +1
Query: 97 MGIISRKIFPACGNMCVCCPALRSSSRQPVKRYKKLLADIFPKSPDELPSERKIVKLCEY 276
MG ISR +FPAC +MC+CCPALRS SRQPVKRYKKLL +IFPKSPD P+ERKIVKLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 277 AAKNPFRIPKIAKYLEERCYKELRSGHIKLVTIITESFNKLLSICKVQIAYFALDVLNVI 456
AAKNP RIPKIAK+LEERCYK+LRS +K + I+TE++NK+L CK Q+AYFA +LNV+
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 457 TELLGYSKDETIQTLGCQSLTRFIFCQVDTTYTHNIEKLVRKVCILSREHGETHEKRC 630
TELL SK +T LGCQ+LTRFI+ QVD TYTH+IEK KVC L+RE GE H+K+C
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQC 178
>gb|AAK56279.1|AF367291_1 At1g05960/T21E18_20 [Arabidopsis thaliana]
gi|25090223|gb|AAN72256.1| At1g05960/T21E18_20
[Arabidopsis thaliana]
Length = 731
Score = 191 bits (484), Expect = 9e-48
Identities = 86/172 (50%), Positives = 128/172 (74%)
Frame = +1
Query: 97 MGIISRKIFPACGNMCVCCPALRSSSRQPVKRYKKLLADIFPKSPDELPSERKIVKLCEY 276
MG++SR++ PACGN+C CP+LR+ SR PVKRYKK+LA+IFP++ + P++RKI KLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 277 AAKNPFRIPKIAKYLEERCYKELRSGHIKLVTIITESFNKLLSICKVQIAYFALDVLNVI 456
A++NP RIPKI +YLE++CYKELR+G+I V ++ + KLLS CK Q+ F+ +L+++
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 457 TELLGYSKDETIQTLGCQSLTRFIFCQVDTTYTHNIEKLVRKVCILSREHGE 612
LL +K+E +Q LGC +L FI Q ++ N+E L+ K+C L++E G+
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGD 172
>pir||F86194 hypothetical protein [imported] - Arabidopsis thaliana
gi|8810459|gb|AAF80120.1|AC024174_2 Contains similarity
to an unknown protein T11A7.7 gi|2335096 from
Arabidopsis thaliana BAC T11A7 gb|AC002339 and contains
a tropomyosin PF|00261 domain. ESTs gb|AI995205,
gb|N37925, gb|F13889, gb|AV523107, gb|AV535948,
gb|AV558461, gb|F13888 come from this gene
Length = 1628
Score = 191 bits (484), Expect = 9e-48
Identities = 86/172 (50%), Positives = 128/172 (74%)
Frame = +1
Query: 97 MGIISRKIFPACGNMCVCCPALRSSSRQPVKRYKKLLADIFPKSPDELPSERKIVKLCEY 276
MG++SR++ PACGN+C CP+LR+ SR PVKRYKK+LA+IFP++ + P++RKI KLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 277 AAKNPFRIPKIAKYLEERCYKELRSGHIKLVTIITESFNKLLSICKVQIAYFALDVLNVI 456
A++NP RIPKI +YLE++CYKELR+G+I V ++ + KLLS CK Q+ F+ +L+++
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 457 TELLGYSKDETIQTLGCQSLTRFIFCQVDTTYTHNIEKLVRKVCILSREHGE 612
LL +K+E +Q LGC +L FI Q ++ N+E L+ K+C L++E G+
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGD 172
>ref|NP_563755.1| expressed protein; protein id: At1g05960.1, supported by cDNA:
gi_14194168 [Arabidopsis thaliana]
Length = 982
Score = 191 bits (484), Expect = 9e-48
Identities = 86/172 (50%), Positives = 128/172 (74%)
Frame = +1
Query: 97 MGIISRKIFPACGNMCVCCPALRSSSRQPVKRYKKLLADIFPKSPDELPSERKIVKLCEY 276
MG++SR++ PACGN+C CP+LR+ SR PVKRYKK+LA+IFP++ + P++RKI KLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 277 AAKNPFRIPKIAKYLEERCYKELRSGHIKLVTIITESFNKLLSICKVQIAYFALDVLNVI 456
A++NP RIPKI +YLE++CYKELR+G+I V ++ + KLLS CK Q+ F+ +L+++
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 457 TELLGYSKDETIQTLGCQSLTRFIFCQVDTTYTHNIEKLVRKVCILSREHGE 612
LL +K+E +Q LGC +L FI Q ++ N+E L+ K+C L++E G+
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGD 172
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 558,452,859
Number of Sequences: 1393205
Number of extensions: 12221376
Number of successful extensions: 31903
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 30786
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31898
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)