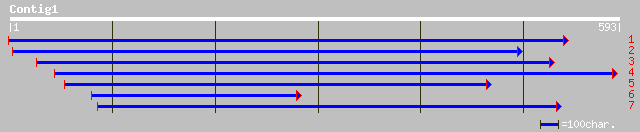
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005429A_C01 KMC005429A_c01
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL15313.1| AT5g09660/F17I14_150 [Arabidopsis thaliana] 243 2e-63
ref|NP_196528.1| microbody NAD-dependent malate dehydrogenase; p... 243 2e-63
sp|Q43743|MDHG_BRANA Malate dehydrogenase 1, glyoxysomal precurs... 242 3e-63
sp|Q9XFW3|MDHH_BRANA Malate dehydrogenase 2, glyoxysomal precurs... 242 3e-63
sp|P46488|MDHG_CUCSA Malate dehydrogenase, glyoxysomal precursor... 238 3e-62
>gb|AAL15313.1| AT5g09660/F17I14_150 [Arabidopsis thaliana]
Length = 354
Score = 243 bits (619), Expect = 2e-63
Identities = 121/138 (87%), Positives = 130/138 (93%)
Frame = -2
Query: 585 GGHAGVTILPLLSQVKPPSSFTAEETDYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 406
GGHAGVTILPLLSQVKPPSSFT +E +YLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK
Sbjct: 216 GGHAGVTILPLLSQVKPPSSFTPQEIEYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 275
Query: 405 FANSCLRGLKGEAGVVECSFVESQVTELPFFATKVRLGRAGAEEIYQLGPLNEYERIGLE 226
FA++CLRGL+G+A VVECSFV SQVTEL FFATKVRLGR GAEE+YQLGPLNEYERIGLE
Sbjct: 276 FADACLRGLRGDANVVECSFVASQVTELAFFATKVRLGRTGAEEVYQLGPLNEYERIGLE 335
Query: 225 KAKRELVGSIQKGIDFIR 172
KAK EL GSIQKG++FIR
Sbjct: 336 KAKDELAGSIQKGVEFIR 353
>ref|NP_196528.1| microbody NAD-dependent malate dehydrogenase; protein id:
At5g09660.1, supported by cDNA: gi_14335145, supported
by cDNA: gi_16226936, supported by cDNA: gi_16323156,
supported by cDNA: gi_18655350 [Arabidopsis thaliana]
gi|11133713|sp|Q9ZP05|MDHG_ARATH Malate dehydrogenase,
glyoxysomal precursor (mbNAD-MDH)
gi|11251113|pir||T49932 malate dehydrogenase (EC
1.1.1.37) precursor, NAD-dependent, glyoxysomal
[validated] - Arabidopsis thaliana
gi|3929651|emb|CAA10321.1| microbody NAD-dependent
malate dehydrogenase [Arabidopsis thaliana]
gi|7671423|emb|CAB89364.1| microbody NAD-dependent
malate dehydrogenase [Arabidopsis thaliana]
gi|9758994|dbj|BAB09521.1| microbody NAD-dependent
malate dehydrogenase [Arabidopsis thaliana]
gi|14335146|gb|AAK59853.1| AT5g09660/F17I14_150
[Arabidopsis thaliana]
gi|16226937|gb|AAL16303.1|AF428373_1
AT5g09660/F17I14_150 [Arabidopsis thaliana]
gi|18655351|gb|AAL76131.1| AT5g09660/F17I14_150
[Arabidopsis thaliana]
Length = 354
Score = 243 bits (619), Expect = 2e-63
Identities = 121/138 (87%), Positives = 130/138 (93%)
Frame = -2
Query: 585 GGHAGVTILPLLSQVKPPSSFTAEETDYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 406
GGHAGVTILPLLSQVKPPSSFT +E +YLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK
Sbjct: 216 GGHAGVTILPLLSQVKPPSSFTPQEIEYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 275
Query: 405 FANSCLRGLKGEAGVVECSFVESQVTELPFFATKVRLGRAGAEEIYQLGPLNEYERIGLE 226
FA++CLRGL+G+A VVECSFV SQVTEL FFATKVRLGR GAEE+YQLGPLNEYERIGLE
Sbjct: 276 FADACLRGLRGDANVVECSFVASQVTELAFFATKVRLGRTGAEEVYQLGPLNEYERIGLE 335
Query: 225 KAKRELVGSIQKGIDFIR 172
KAK EL GSIQKG++FIR
Sbjct: 336 KAKDELAGSIQKGVEFIR 353
>sp|Q43743|MDHG_BRANA Malate dehydrogenase 1, glyoxysomal precursor
gi|4995089|emb|CAB43994.1| malate dehydrogenase 1
[Brassica napus]
Length = 358
Score = 242 bits (617), Expect = 3e-63
Identities = 119/138 (86%), Positives = 129/138 (93%)
Frame = -2
Query: 585 GGHAGVTILPLLSQVKPPSSFTAEETDYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 406
GGHAGVTILPLLSQVKPPSSFT E +YLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK
Sbjct: 220 GGHAGVTILPLLSQVKPPSSFTPSEIEYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 279
Query: 405 FANSCLRGLKGEAGVVECSFVESQVTELPFFATKVRLGRAGAEEIYQLGPLNEYERIGLE 226
FA++CLRGL+G+A V+ECSFV SQVTEL FFATKVRLGR GAEE++QLGPLNEYER+GLE
Sbjct: 280 FADACLRGLRGDANVIECSFVASQVTELAFFATKVRLGRTGAEEVFQLGPLNEYERVGLE 339
Query: 225 KAKRELVGSIQKGIDFIR 172
KAK EL GSIQKG+DFIR
Sbjct: 340 KAKEELAGSIQKGVDFIR 357
>sp|Q9XFW3|MDHH_BRANA Malate dehydrogenase 2, glyoxysomal precursor
gi|4995091|emb|CAB43995.1| malate dehydrogenase 2
[Brassica napus]
Length = 358
Score = 242 bits (617), Expect = 3e-63
Identities = 119/138 (86%), Positives = 129/138 (93%)
Frame = -2
Query: 585 GGHAGVTILPLLSQVKPPSSFTAEETDYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 406
GGHAGVTILPLLSQVKPPSSFT E +YLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK
Sbjct: 220 GGHAGVTILPLLSQVKPPSSFTPSEIEYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 279
Query: 405 FANSCLRGLKGEAGVVECSFVESQVTELPFFATKVRLGRAGAEEIYQLGPLNEYERIGLE 226
FA++CLRGL+G+A V+ECSFV SQVTEL FFATKVRLGR GAEE++QLGPLNEYER+GLE
Sbjct: 280 FADACLRGLRGDANVIECSFVASQVTELAFFATKVRLGRTGAEEVFQLGPLNEYERVGLE 339
Query: 225 KAKRELVGSIQKGIDFIR 172
KAK EL GSIQKG+DFIR
Sbjct: 340 KAKEELAGSIQKGVDFIR 357
>sp|P46488|MDHG_CUCSA Malate dehydrogenase, glyoxysomal precursor gi|1076276|pir||S52039
malate dehydrogenase (EC 1.1.1.37) - cucumber
gi|695311|gb|AAC41647.1| glyoxysomal malate
dehydrogenase
Length = 356
Score = 238 bits (608), Expect = 3e-62
Identities = 117/138 (84%), Positives = 129/138 (92%)
Frame = -2
Query: 585 GGHAGVTILPLLSQVKPPSSFTAEETDYLTNRIQNGGTEVVEAKAGAGSATLSMAYAAAK 406
GGHAGVTILPLLSQVKPPSSFT EE +YLT+RIQNGGTEVVEAKAGAGSATLSMAYAA K
Sbjct: 218 GGHAGVTILPLLSQVKPPSSFTQEEINYLTDRIQNGGTEVVEAKAGAGSATLSMAYAAVK 277
Query: 405 FANSCLRGLKGEAGVVECSFVESQVTELPFFATKVRLGRAGAEEIYQLGPLNEYERIGLE 226
FA++CLRGL+G+AGVVEC+FV SQVTELPFFATKVRLGR G +E+Y LGPLNEYERIGLE
Sbjct: 278 FADACLRGLRGDAGVVECAFVSSQVTELPFFATKVRLGRNGIDEVYSLGPLNEYERIGLE 337
Query: 225 KAKRELVGSIQKGIDFIR 172
KAK+EL GSI+KG+ FIR
Sbjct: 338 KAKKELAGSIEKGVSFIR 355
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 506,318,898
Number of Sequences: 1393205
Number of extensions: 11165854
Number of successful extensions: 32178
Number of sequences better than 10.0: 259
Number of HSP's better than 10.0 without gapping: 30866
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31966
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)