Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
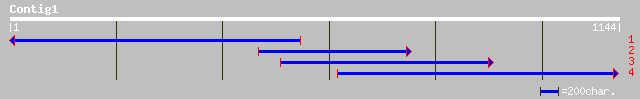
Query= KMC005413A_C01 KMC005413A_c01
(1144 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_567851.1| putative protein; protein id: At4g30690.1, supp... 162 9e-39
ref|NP_179984.1| putative chloroplast initiation factor 3; prote... 154 3e-36
ref|NP_623290.1| Translation initiation factor IF3 [Thermoanaero... 77 7e-13
sp|P36177|IF3C_EUGGR TRANSLATION INITIATION FACTOR IF-3, CHLOROP... 77 7e-13
ref|NP_488663.1| translation initiation factor IF-3 [Nostoc sp. ... 75 1e-12
>ref|NP_567851.1| putative protein; protein id: At4g30690.1, supported by cDNA:
gi_14596120, supported by cDNA: gi_18377497 [Arabidopsis
thaliana] gi|14596121|gb|AAK68788.1| putative protein
[Arabidopsis thaliana] gi|18377498|gb|AAL66915.1|
putative protein [Arabidopsis thaliana]
Length = 281
Score = 162 bits (410), Expect = 9e-39
Identities = 77/116 (66%), Positives = 94/116 (80%)
Frame = -3
Query: 1142 MDLKELKMGYNIDQHDYSVRLRAARKFLTDGDKVKVIVNLKGRENEFRNIAIELIKRFQN 963
MDLKELKMGYNIDQHDYSVR+RAARKFL DGDKVKVIVN+KGRENEFRNIAIEL++RFQ
Sbjct: 163 MDLKELKMGYNIDQHDYSVRMRAARKFLQDGDKVKVIVNMKGRENEFRNIAIELLRRFQT 222
Query: 962 DVGELGTEEAKSFRDRNIFILLVPESNCYTENSRTTPRKKTSQQPDEVTVDEVSVS 795
++GELGTEE+K+FRDRN+FI+LVP + P+KK D+V+ ++ +
Sbjct: 223 EIGELGTEESKNFRDRNLFIVLVPNKEVIRKVQEPPPKKKKKPADDKVSAANITAT 278
>ref|NP_179984.1| putative chloroplast initiation factor 3; protein id: At2g24060.1,
supported by cDNA: 41558., supported by cDNA:
gi_18176091, supported by cDNA: gi_20465300 [Arabidopsis
thaliana] gi|25412188|pir||B84632 probable chloroplast
initiation factor 3 [imported] - Arabidopsis thaliana
gi|3738333|gb|AAC63674.1| putative chloroplast initiation
factor 3 [Arabidopsis thaliana]
gi|18176092|gb|AAL59982.1| putative chloroplast
initiation factor 3 [Arabidopsis thaliana]
gi|20465301|gb|AAM20054.1| putative chloroplast
initiation factor 3 [Arabidopsis thaliana]
gi|21593709|gb|AAM65676.1| putative chloroplast
initiation factor 3 [Arabidopsis thaliana]
Length = 312
Score = 154 bits (388), Expect = 3e-36
Identities = 74/116 (63%), Positives = 90/116 (76%)
Frame = -3
Query: 1142 MDLKELKMGYNIDQHDYSVRLRAARKFLTDGDKVKVIVNLKGRENEFRNIAIELIKRFQN 963
MDLKELKMGYNIDQHDYSVRLRAA+KFL DGDKVKVIV++KGRENEFRNIAIEL++RFQ
Sbjct: 167 MDLKELKMGYNIDQHDYSVRLRAAQKFLQDGDKVKVIVSMKGRENEFRNIAIELLRRFQT 226
Query: 962 DVGELGTEEAKSFRDRNIFILLVPESNCYTENSRTTPRKKTSQQPDEVTVDEVSVS 795
++GEL TEE+K+FRDRN+FI+LVP + RKK +E + ++
Sbjct: 227 EIGELATEESKNFRDRNMFIILVPNKEMIRKPQEPPTRKKKKTAENEASASAAEIT 282
>ref|NP_623290.1| Translation initiation factor IF3 [Thermoanaerobacter tengcongensis]
gi|22095768|sp|Q8R9C2|IF3_THETN Translation initiation
factor IF-3 gi|20516706|gb|AAM24894.1| Translation
initiation factor IF3 [Thermoanaerobacter tengcongensis]
Length = 180
Score = 76.6 bits (187), Expect = 7e-13
Identities = 36/86 (41%), Positives = 60/86 (68%), Gaps = 1/86 (1%)
Frame = -3
Query: 1142 MDLKELKMGYNIDQHDYSVRLRAARKFLTDGDKVKVIVNLKGRENEFRNIAIELIKRFQN 963
+++KE++M NI+ HD+ V+L++A KFL DGDKVKV + +GRE ++A +L+KRF
Sbjct: 93 INVKEIRMSPNIEDHDFGVKLKSAIKFLKDGDKVKVTIRFRGREAAHTSLAEDLLKRFAE 152
Query: 962 DVGELG-TEEAKSFRDRNIFILLVPE 888
++ E G E+A S RN+ +++ P+
Sbjct: 153 ELREYGNVEKAPSMDGRNMMMVIAPK 178
>sp|P36177|IF3C_EUGGR TRANSLATION INITIATION FACTOR IF-3, CHLOROPLAST PRECURSOR (IF-3CHL)
gi|7484345|pir||A54391 translation initiation factor IF-3
precursor, chloroplast - Euglena gracilis
gi|475786|gb|AAA20996.1| chloroplast initiation factor 3
Length = 538
Score = 76.6 bits (187), Expect = 7e-13
Identities = 34/69 (49%), Positives = 50/69 (72%)
Frame = -3
Query: 1139 DLKELKMGYNIDQHDYSVRLRAARKFLTDGDKVKVIVNLKGRENEFRNIAIELIKRFQND 960
++KELK+ + I QHDY VR++ ARKFL G ++KV + KGREN+F I ++KRFQND
Sbjct: 388 EVKELKVSHKIGQHDYDVRVKQARKFLEGGHRIKVSMEFKGRENQFVEIGRAVMKRFQND 447
Query: 959 VGELGTEEA 933
+ ++G +A
Sbjct: 448 LADMGKADA 456
>ref|NP_488663.1| translation initiation factor IF-3 [Nostoc sp. PCC 7120]
gi|25299585|pir||AG2383 translation initiation factor
IF-3 [imported] - Nostoc sp. (strain PCC 7120)
gi|17133760|dbj|BAB76322.1| translation initiation factor
IF-3 [Nostoc sp. PCC 7120]
Length = 204
Score = 75.5 bits (184), Expect = 1e-12
Identities = 36/85 (42%), Positives = 57/85 (66%), Gaps = 1/85 (1%)
Frame = -3
Query: 1139 DLKELKMGYNIDQHDYSVRLRAARKFLTDGDKVKVIVNLKGRENEFRNIAIELIKRFQND 960
D+KE+KM Y I++HDY+VR++ A +FL DGDKVK V +GRE + ++A +L+KR D
Sbjct: 119 DVKEVKMRYKIEEHDYNVRVKQAERFLKDGDKVKATVMFRGREIQHSDLAEDLLKRMATD 178
Query: 959 VGELG-TEEAKSFRDRNIFILLVPE 888
+ G ++A RN+ +L+ P+
Sbjct: 179 LEPFGEVQQAPKKEGRNMMMLISPK 203
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 898,804,534
Number of Sequences: 1393205
Number of extensions: 19102883
Number of successful extensions: 44512
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 41464
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44374
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 70007348610
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)