Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005412A_C01 KMC005412A_c01
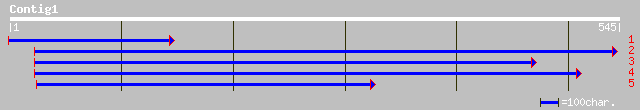
(520 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S51590 mitochondrial processing peptidase (EC 3.4.24.64) al... 139 2e-32
dbj|BAB55500.1| putative mitochondrial processing peptidase alph... 137 6e-32
gb|AAK59675.1| putative mitochondrial processing peptidase alpha... 137 1e-31
ref|NP_566548.1| putative mitochondrial processing peptidase al... 137 1e-31
sp|P29677|MPPA_SOLTU Mitochondrial processing peptidase alpha su... 136 1e-31
>pir||S51590 mitochondrial processing peptidase (EC 3.4.24.64) alpha-II chain
precursor - potato gi|587562|emb|CAA56520.1|
mitochondrial processing peptidase [Solanum tuberosum]
Length = 504
Score = 139 bits (350), Expect = 2e-32
Identities = 67/96 (69%), Positives = 84/96 (86%)
Frame = -2
Query: 519 KVSQDQLDRAKKSTKSAVLMNLESRMIASEDIGRQILTYGERKPVEQFLKAVDAITLNDI 340
+V QLDRAK+STKSA+LMNLESRM+ASEDIGRQ+L YGERKPVE LKA+DAI+ NDI
Sbjct: 409 EVDMVQLDRAKQSTKSAILMNLESRMVASEDIGRQLLIYGERKPVEHVLKAIDAISANDI 468
Query: 339 TKISQKIISSPLTMASYGDVVNVPSYESVNRIFHAK 232
++QK+ISSPLTMASYGDV+++P+Y+ V+ FH+K
Sbjct: 469 ASVAQKLISSPLTMASYGDVLSLPTYDVVSSRFHSK 504
>dbj|BAB55500.1| putative mitochondrial processing peptidase alpha subuunit,
mitochondrial recursor(ALPHA-MPP) [Oryza sativa
(japonica cultivar-group)]
Length = 884
Score = 137 bits (346), Expect = 6e-32
Identities = 70/98 (71%), Positives = 88/98 (89%), Gaps = 5/98 (5%)
Frame = -2
Query: 519 KVSQDQLDRAKKSTKSAVLMNLESRMIASEDIGRQILTYGERKPVEQFLKAVDAITLNDI 340
KV+Q+QLDRAK++TKSAVLMNLESR++ASEDIGRQILTYGERKP+E FLK ++AITLNDI
Sbjct: 456 KVTQEQLDRAKQATKSAVLMNLESRVVASEDIGRQILTYGERKPIEHFLKDLEAITLNDI 515
Query: 339 TKISQKIISSPLTMASYGD-----VVNVPSYESVNRIF 241
+ ++KIISSPLT+AS+GD V++VPSYESV++ F
Sbjct: 516 SSTAKKIISSPLTLASWGDECFLAVIHVPSYESVSQKF 553
>gb|AAK59675.1| putative mitochondrial processing peptidase alpha subunit
[Arabidopsis thaliana]
Length = 499
Score = 137 bits (344), Expect = 1e-31
Identities = 66/93 (70%), Positives = 81/93 (86%)
Frame = -2
Query: 519 KVSQDQLDRAKKSTKSAVLMNLESRMIASEDIGRQILTYGERKPVEQFLKAVDAITLNDI 340
KV+Q LDRAK +TKSA+LMNLESRMIA+EDIGRQILTYGERKPV+QFLK VD +TL DI
Sbjct: 406 KVNQKHLDRAKAATKSAILMNLESRMIAAEDIGRQILTYGERKPVDQFLKTVDQLTLKDI 465
Query: 339 TKISQKIISSPLTMASYGDVVNVPSYESVNRIF 241
+ K+I+ PLTMA++GDV+NVPSY+SV++ F
Sbjct: 466 ADFTSKVITKPLTMATFGDVLNVPSYDSVSKRF 498
>ref|NP_566548.1| putative mitochondrial processing peptidase alpha subunit; protein
id: At3g16480.1, supported by cDNA: gi_14334533
[Arabidopsis thaliana] gi|2062155|gb|AAB63629.1|
mitochondrial processing peptidase alpha subunit
precusor isolog [Arabidopsis thaliana]
gi|9279647|dbj|BAB01147.1| mitochondrial processing
peptidase alpha subunit [Arabidopsis thaliana]
gi|23297133|gb|AAN13101.1| putative mitochondrial
processing peptidase alpha subunit [Arabidopsis
thaliana]
Length = 499
Score = 137 bits (344), Expect = 1e-31
Identities = 66/93 (70%), Positives = 81/93 (86%)
Frame = -2
Query: 519 KVSQDQLDRAKKSTKSAVLMNLESRMIASEDIGRQILTYGERKPVEQFLKAVDAITLNDI 340
KV+Q LDRAK +TKSA+LMNLESRMIA+EDIGRQILTYGERKPV+QFLK VD +TL DI
Sbjct: 406 KVNQKHLDRAKAATKSAILMNLESRMIAAEDIGRQILTYGERKPVDQFLKTVDQLTLKDI 465
Query: 339 TKISQKIISSPLTMASYGDVVNVPSYESVNRIF 241
+ K+I+ PLTMA++GDV+NVPSY+SV++ F
Sbjct: 466 ADFTSKVITKPLTMATFGDVLNVPSYDSVSKRF 498
>sp|P29677|MPPA_SOLTU Mitochondrial processing peptidase alpha subunit, mitochondrial
precursor (Alpha-MPP) (Ubiquinol-cytochrome C reductase
subunit II) gi|421956|pir||S23558 ubiquinol-cytochrome-c
reductase (EC 1.10.2.2) alpha chain precursor - potato
gi|21493|emb|CAA46990.1| mitochondrial processing
peptidase [Solanum tuberosum]
Length = 504
Score = 136 bits (343), Expect = 1e-31
Identities = 65/96 (67%), Positives = 84/96 (86%)
Frame = -2
Query: 519 KVSQDQLDRAKKSTKSAVLMNLESRMIASEDIGRQILTYGERKPVEQFLKAVDAITLNDI 340
+V Q QL+RAK++TKSA+LMNLESRM+ASEDIGRQ+LTYGER PVE FLKA+DA++ DI
Sbjct: 409 EVDQVQLNRAKQATKSAILMNLESRMVASEDIGRQLLTYGERNPVEHFLKAIDAVSAKDI 468
Query: 339 TKISQKIISSPLTMASYGDVVNVPSYESVNRIFHAK 232
+ QK+ISSPLTMASYGDV+++PSY++V+ F +K
Sbjct: 469 ASVVQKLISSPLTMASYGDVLSLPSYDAVSSRFRSK 504
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 435,335,688
Number of Sequences: 1393205
Number of extensions: 8919619
Number of successful extensions: 20127
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 19586
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20104
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16442828304
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)