Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005402A_C01 KMC005402A_c01
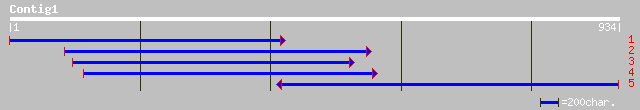
(929 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_567639.1| Expressed protein; protein id: At4g21860.1, sup... 256 3e-67
gb|AAM62876.1| unknown [Arabidopsis thaliana] 254 1e-66
gb|AAO72582.1| unknown [Oryza sativa (japonica cultivar-group)] 239 6e-62
ref|NP_567271.1| putative protein; protein id: At4g04800.1, supp... 235 8e-61
gb|AAD03449.1| contains similarity to Methanobacterium thermoaut... 234 1e-60
>ref|NP_567639.1| Expressed protein; protein id: At4g21860.1, supported by cDNA:
16643., supported by cDNA: gi_13507550, supported by
cDNA: gi_15293288 [Arabidopsis thaliana]
gi|13507551|gb|AAK28638.1|AF360341_1 unknown protein
[Arabidopsis thaliana] gi|15293289|gb|AAK93755.1|
unknown protein [Arabidopsis thaliana]
Length = 202
Score = 256 bits (655), Expect = 3e-67
Identities = 124/174 (71%), Positives = 140/174 (80%), Gaps = 5/174 (2%)
Frame = -1
Query: 926 SHSTSHFTVFPSNPIITPSSISIRHQQRKRGFRGGVVVAM--AAPQ---KSEDEWRAVLS 762
S S + F S+P+ PS +RGF GG +VAM +AP+ K E+EWRA+LS
Sbjct: 34 SPSRRNLLRFSSSPLSFPSL--------RRGFHGGRIVAMGSSAPESVNKPEEEWRAILS 85
Query: 761 PEQFRILRQKGTEYPGTGEYDKFFAEGVYRCAGCETPLYRSTTKFNSGCGWPAFYEGLPG 582
PEQFRILRQKGTEYPGTGEY+K F +G+Y CAGC TPLY+STTKF+SGCGWPAF++GLPG
Sbjct: 86 PEQFRILRQKGTEYPGTGEYNKVFDDGIYCCAGCGTPLYKSTTKFDSGCGWPAFFDGLPG 145
Query: 581 AIYRNPDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERHCVNSISLKFVPAN 420
AI R PDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERHCVNSISLKF P N
Sbjct: 146 AITRTPDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERHCVNSISLKFTPEN 199
>gb|AAM62876.1| unknown [Arabidopsis thaliana]
Length = 202
Score = 254 bits (649), Expect = 1e-66
Identities = 122/174 (70%), Positives = 139/174 (79%), Gaps = 5/174 (2%)
Frame = -1
Query: 926 SHSTSHFTVFPSNPIITPSSISIRHQQRKRGFRGGVVVAM--AAPQ---KSEDEWRAVLS 762
S S + F S+P+ PS +RGF GG +V+M +AP+ K E+EWR +LS
Sbjct: 34 SRSHRNLLRFSSSPLSFPSL--------RRGFHGGRIVSMGSSAPESVNKPEEEWRTILS 85
Query: 761 PEQFRILRQKGTEYPGTGEYDKFFAEGVYRCAGCETPLYRSTTKFNSGCGWPAFYEGLPG 582
PEQFRILRQKGTEYPGTGEY+K F +G+Y CAGC TPLY+STTKF+SGCGWPAF++GLPG
Sbjct: 86 PEQFRILRQKGTEYPGTGEYNKVFDDGIYCCAGCGTPLYKSTTKFDSGCGWPAFFDGLPG 145
Query: 581 AIYRNPDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERHCVNSISLKFVPAN 420
AI R PDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERHCVNSISLKF P N
Sbjct: 146 AITRTPDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERHCVNSISLKFTPEN 199
>gb|AAO72582.1| unknown [Oryza sativa (japonica cultivar-group)]
Length = 168
Score = 239 bits (609), Expect = 6e-62
Identities = 104/126 (82%), Positives = 117/126 (92%)
Frame = -1
Query: 797 QKSEDEWRAVLSPEQFRILRQKGTEYPGTGEYDKFFAEGVYRCAGCETPLYRSTTKFNSG 618
Q+S++EWRAVLSPEQFRILR KGTE PGTGEY+KF+ +GVY CAGC TPLY+STTKF+SG
Sbjct: 43 QRSDEEWRAVLSPEQFRILRLKGTELPGTGEYNKFYGDGVYNCAGCGTPLYKSTTKFDSG 102
Query: 617 CGWPAFYEGLPGAIYRNPDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERHCVNSISL 438
CGWPAF+EGLPGAI R PDPDGRR+EITCAACGGHLGHVFKGEGF TPTDERHCVNS+S+
Sbjct: 103 CGWPAFFEGLPGAINRTPDPDGRRVEITCAACGGHLGHVFKGEGFKTPTDERHCVNSVSI 162
Query: 437 KFVPAN 420
KF PA+
Sbjct: 163 KFTPAS 168
>ref|NP_567271.1| putative protein; protein id: At4g04800.1, supported by cDNA:
39698. [Arabidopsis thaliana]
Length = 176
Score = 235 bits (599), Expect = 8e-61
Identities = 107/134 (79%), Positives = 115/134 (84%), Gaps = 3/134 (2%)
Frame = -1
Query: 815 VAMAAP---QKSEDEWRAVLSPEQFRILRQKGTEYPGTGEYDKFFAEGVYRCAGCETPLY 645
VAMAAP QK ++EWRA+LSPEQFRILRQKGTEYPGTGEY F EGVY C GC PLY
Sbjct: 39 VAMAAPGSVQKGDEEWRAILSPEQFRILRQKGTEYPGTGEYVNFDKEGVYGCVGCNAPLY 98
Query: 644 RSTTKFNSGCGWPAFYEGLPGAIYRNPDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDE 465
+STTKFN+GCGWPAF+EG+PGAI R DPDGRRIEI CA CGGHLGHVFKGEGF TPTDE
Sbjct: 99 KSTTKFNAGCGWPAFFEGIPGAITRTTDPDGRRIEINCATCGGHLGHVFKGEGFATPTDE 158
Query: 464 RHCVNSISLKFVPA 423
RHCVNS+SLKF PA
Sbjct: 159 RHCVNSVSLKFTPA 172
>gb|AAD03449.1| contains similarity to Methanobacterium thermoautotrophicum
transcriptional regulator (GB:AE000850) [Arabidopsis
thaliana]
Length = 281
Score = 234 bits (598), Expect = 1e-60
Identities = 107/137 (78%), Positives = 116/137 (84%), Gaps = 3/137 (2%)
Frame = -1
Query: 809 MAAP---QKSEDEWRAVLSPEQFRILRQKGTEYPGTGEYDKFFAEGVYRCAGCETPLYRS 639
MAAP QK ++EWRA+LSPEQFRILRQKGTEYPGTGEY F EGVY C GC PLY+S
Sbjct: 1 MAAPGSVQKGDEEWRAILSPEQFRILRQKGTEYPGTGEYVNFDKEGVYGCVGCNAPLYKS 60
Query: 638 TTKFNSGCGWPAFYEGLPGAIYRNPDPDGRRIEITCAACGGHLGHVFKGEGFPTPTDERH 459
TTKFN+GCGWPAF+EG+PGAI R DPDGRRIEI CA CGGHLGHVFKGEGF TPTDERH
Sbjct: 61 TTKFNAGCGWPAFFEGIPGAITRTTDPDGRRIEINCATCGGHLGHVFKGEGFATPTDERH 120
Query: 458 CVNSISLKFVPANT*FF 408
CVNS+SLKF PA + FF
Sbjct: 121 CVNSVSLKFTPAASSFF 137
Score = 221 bits (562), Expect = 2e-56
Identities = 102/137 (74%), Positives = 115/137 (83%), Gaps = 4/137 (2%)
Frame = -1
Query: 821 VVVAMA----APQKSEDEWRAVLSPEQFRILRQKGTEYPGTGEYDKFFAEGVYRCAGCET 654
VVV MA +K+E+EWRAVLSPEQFRILRQKGTE PGT EYDKFF EG++ C GC+T
Sbjct: 139 VVVVMADLVTVVKKTEEEWRAVLSPEQFRILRQKGTETPGTEEYDKFFEEGIFSCIGCKT 198
Query: 653 PLYRSTTKFNSGCGWPAFYEGLPGAIYRNPDPDGRRIEITCAACGGHLGHVFKGEGFPTP 474
PLY+STTKF++GCGWPAF+EGLPGAI R PDPDGRR EITCA C GHLGHV KGEG+ TP
Sbjct: 199 PLYKSTTKFDAGCGWPAFFEGLPGAINRAPDPDGRRTEITCAVCDGHLGHVHKGEGYSTP 258
Query: 473 TDERHCVNSISLKFVPA 423
TDER CVNS+S+ F PA
Sbjct: 259 TDERLCVNSVSINFNPA 275
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 833,994,306
Number of Sequences: 1393205
Number of extensions: 19454786
Number of successful extensions: 48776
Number of sequences better than 10.0: 204
Number of HSP's better than 10.0 without gapping: 45997
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48371
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51582455952
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)