Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
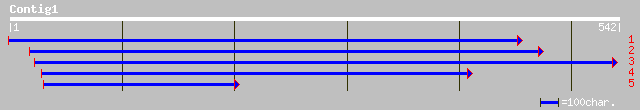
Query= KMC005398A_C01 KMC005398A_c01
(540 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG34815.1|AF243380_1 glutathione S-transferase GST 25 [Glyci... 172 2e-42
sp|P57108|GSTZ_EUPES Glutathione S-transferase zeta class gi|809... 138 4e-32
gb|AAN39918.1| glutathione S-transferase [Capsicum annuum] 132 3e-30
ref|NP_178343.1| glutathione transferase, putative; protein id: ... 125 4e-28
ref|NP_178344.1| glutathione transferase, putative; protein id: ... 121 5e-27
>gb|AAG34815.1|AF243380_1 glutathione S-transferase GST 25 [Glycine max]
Length = 219
Score = 172 bits (437), Expect = 2e-42
Identities = 80/96 (83%), Positives = 88/96 (91%)
Frame = -2
Query: 539 NYVGEKGGPDAKIPWVQSVIRKGFTALEKLLKEHAGRYATGDEVFLADIFLAPQLHAAYN 360
NY+GEK GPD K+PW QS+IR+GF ALEKLLK+H GRYATGDEVFLADIFLAPQLHAA+
Sbjct: 122 NYIGEKVGPDEKLPWAQSIIRRGFKALEKLLKDHTGRYATGDEVFLADIFLAPQLHAAFK 181
Query: 359 RFNIQMDEFPILARLHETYYEIPAFREALPENQPDA 252
RFNI M+EFPILARLHETY EIPAF+EALPENQPDA
Sbjct: 182 RFNIHMNEFPILARLHETYNEIPAFQEALPENQPDA 217
>sp|P57108|GSTZ_EUPES Glutathione S-transferase zeta class
gi|8099671|gb|AAF72197.1|AF263737_1 glutathione
S-transferase [Euphorbia esula]
Length = 225
Score = 138 bits (348), Expect = 4e-32
Identities = 60/96 (62%), Positives = 77/96 (79%)
Frame = -2
Query: 539 NYVGEKGGPDAKIPWVQSVIRKGFTALEKLLKEHAGRYATGDEVFLADIFLAPQLHAAYN 360
N++GEK PD K+PWVQ I KGF ALEKLL+ HAGR+ATGDEV+LAD+FL PQ+HAA
Sbjct: 124 NFIGEKVSPDEKVPWVQRHISKGFAALEKLLQGHAGRFATGDEVYLADLFLEPQIHAAIT 183
Query: 359 RFNIQMDEFPILARLHETYYEIPAFREALPENQPDA 252
RFN+ M +FP+L RLHE Y ++P F+ A+P+ QPD+
Sbjct: 184 RFNVDMTQFPLLLRLHEAYSQLPEFQNAMPDKQPDS 219
>gb|AAN39918.1| glutathione S-transferase [Capsicum annuum]
Length = 220
Score = 132 bits (332), Expect = 3e-30
Identities = 58/95 (61%), Positives = 78/95 (82%)
Frame = -2
Query: 536 YVGEKGGPDAKIPWVQSVIRKGFTALEKLLKEHAGRYATGDEVFLADIFLAPQLHAAYNR 357
Y+ EK GP+ PW QS I KGF ALEKLLK++AG+YATGDEV++AD+FLAPQ++AA NR
Sbjct: 124 YIQEKVGPNETTPWAQSHITKGFEALEKLLKDYAGKYATGDEVYMADLFLAPQIYAAINR 183
Query: 356 FNIQMDEFPILARLHETYYEIPAFREALPENQPDA 252
F + M++FP L R+++ Y E+PAF++A+PE QPDA
Sbjct: 184 FEVDMNQFPTLLRVYKAYQELPAFQDAMPEKQPDA 218
>ref|NP_178343.1| glutathione transferase, putative; protein id: At2g02380.1
[Arabidopsis thaliana] gi|11133278|sp|Q9ZVQ4|GTZ2_ARATH
Probable glutathione S-transferase zeta-class 2
gi|25317409|pir||A84436 probable glutathione
S-transferase [imported] - Arabidopsis thaliana
gi|3894170|gb|AAC78520.1| putative glutathione
S-transferase [Arabidopsis thaliana]
Length = 223
Score = 125 bits (313), Expect = 4e-28
Identities = 57/94 (60%), Positives = 74/94 (78%)
Frame = -2
Query: 536 YVGEKGGPDAKIPWVQSVIRKGFTALEKLLKEHAGRYATGDEVFLADIFLAPQLHAAYNR 357
Y+ +K + K W+ + I KGFTALEKLL AG+YATGDEV+LAD+FLAPQ+HAA+NR
Sbjct: 125 YLEDKINAEEKTAWITNAITKGFTALEKLLVSCAGKYATGDEVYLADLFLAPQIHAAFNR 184
Query: 356 FNIQMDEFPILARLHETYYEIPAFREALPENQPD 255
F+I M+ FP LAR +E+Y E+PAF+ A+PE QPD
Sbjct: 185 FHINMEPFPTLARFYESYNELPAFQNAVPEKQPD 218
>ref|NP_178344.1| glutathione transferase, putative; protein id: At2g02390.1,
supported by cDNA: gi_11095997, supported by cDNA:
gi_15450462, supported by cDNA: gi_16974448 [Arabidopsis
thaliana] gi|11133276|sp|Q9ZVQ3|GTZ1_ARATH Glutathione
S-transferase zeta-class 1 (AtGSTZ1) (Maleylacetone
isomerase) (MAI) gi|25317411|pir||B84436 probable
glutathione S-transferase [imported] - Arabidopsis
thaliana gi|14719799|pdb|1E6B|A Chain A, Crystal
Structure Of A Zeta Class Glutathione S-Transferase From
Arabidopsis Thaliana gi|3894171|gb|AAC78521.1| putative
glutathione S-transferase [Arabidopsis thaliana]
gi|11095998|gb|AAG30131.1|AF288182_1 glutathione
S-transferase [Arabidopsis thaliana]
gi|11967659|emb|CAC19475.1| glutathione transferase zeta
1 [Arabidopsis thaliana] gi|15450463|gb|AAK96525.1|
At2g02390/T16F16.18 [Arabidopsis thaliana]
gi|16974449|gb|AAL31228.1| At2g02390/T16F16.18
[Arabidopsis thaliana] gi|28932692|gb|AAO60039.1|
glutathione S-transferase zeta [Arabidopsis thaliana]
Length = 221
Score = 121 bits (304), Expect = 5e-27
Identities = 56/95 (58%), Positives = 72/95 (74%)
Frame = -2
Query: 536 YVGEKGGPDAKIPWVQSVIRKGFTALEKLLKEHAGRYATGDEVFLADIFLAPQLHAAYNR 357
Y+ EK + K WV + I KGFTALEKLL AG++ATGDE++LAD+FLAPQ+H A NR
Sbjct: 122 YIEEKINVEEKTAWVNNAITKGFTALEKLLVNCAGKHATGDEIYLADLFLAPQIHGAINR 181
Query: 356 FNIQMDEFPILARLHETYYEIPAFREALPENQPDA 252
F I M+ +P LA+ +E+Y E+PAF+ ALPE QPDA
Sbjct: 182 FQINMEPYPTLAKCYESYNELPAFQNALPEKQPDA 216
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,828,020
Number of Sequences: 1393205
Number of extensions: 10283908
Number of successful extensions: 19587
Number of sequences better than 10.0: 149
Number of HSP's better than 10.0 without gapping: 19217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19549
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)