Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
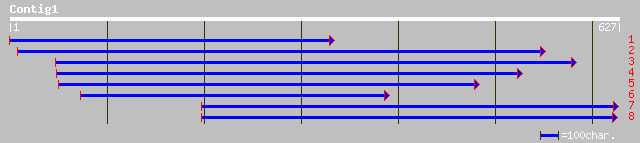
Query= KMC005389A_C01 KMC005389A_c01
(624 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK59627.1| unknown protein [Arabidopsis thaliana] 166 2e-40
ref|NP_563902.1| chloroplast lumen pentapeptide protein, putativ... 166 2e-40
pir||F86257 Hypothetical protein [imported] - Arabidopsis thalia... 151 8e-36
ref|NP_443033.1| unknown protein [Synechocystis sp. PCC 6803] gi... 64 2e-09
ref|ZP_00115441.1| hypothetical protein [Synechococcus sp. WH 8102] 61 1e-08
>gb|AAK59627.1| unknown protein [Arabidopsis thaliana]
Length = 280
Score = 166 bits (421), Expect = 2e-40
Identities = 79/95 (83%), Positives = 87/95 (91%)
Frame = -3
Query: 622 AILVRTVLTRSDLGGSIIEGADFSDAVLDLTQKLALCKYASGTNPVTGVSTRVSLGCGNK 443
A+LVR+VLTRSDLGG+ IEGADFSDAV+DL QK ALCKYA+GTNP+TGV TR SLGCGN
Sbjct: 184 AVLVRSVLTRSDLGGAKIEGADFSDAVIDLLQKQALCKYATGTNPLTGVDTRKSLGCGNS 243
Query: 442 RRNAYGTPSSPLLSAPPQKLLNRDGFCDEATGLCD 338
RRNAYG+PSSPLLSAPPQ+LL RDGFCDE TGLCD
Sbjct: 244 RRNAYGSPSSPLLSAPPQRLLGRDGFCDEKTGLCD 278
>ref|NP_563902.1| chloroplast lumen pentapeptide protein, putative; protein id:
At1g12250.1, supported by cDNA: gi_14334897 [Arabidopsis
thaliana] gi|23297125|gb|AAN13098.1| unknown protein
[Arabidopsis thaliana]
Length = 280
Score = 166 bits (421), Expect = 2e-40
Identities = 79/95 (83%), Positives = 87/95 (91%)
Frame = -3
Query: 622 AILVRTVLTRSDLGGSIIEGADFSDAVLDLTQKLALCKYASGTNPVTGVSTRVSLGCGNK 443
A+LVR+VLTRSDLGG+ IEGADFSDAV+DL QK ALCKYA+GTNP+TGV TR SLGCGN
Sbjct: 184 AVLVRSVLTRSDLGGAKIEGADFSDAVIDLLQKQALCKYATGTNPLTGVDTRKSLGCGNS 243
Query: 442 RRNAYGTPSSPLLSAPPQKLLNRDGFCDEATGLCD 338
RRNAYG+PSSPLLSAPPQ+LL RDGFCDE TGLCD
Sbjct: 244 RRNAYGSPSSPLLSAPPQRLLGRDGFCDEKTGLCD 278
>pir||F86257 Hypothetical protein [imported] - Arabidopsis thaliana
gi|10086510|gb|AAG12570.1|AC022522_3 Hypothetical
protein [Arabidopsis thaliana]
Length = 293
Score = 151 bits (381), Expect = 8e-36
Identities = 79/124 (63%), Positives = 87/124 (69%), Gaps = 29/124 (23%)
Frame = -3
Query: 622 AILVRTVLTRSDLGGSIIEGADFSDAVLDLTQKL-------------------------- 521
A+LVR+VLTRSDLGG+ IEGADFSDAV+DL QK
Sbjct: 168 AVLVRSVLTRSDLGGAKIEGADFSDAVIDLLQKQVTTTHHYIYPSFRSTIKKYFTNGFHN 227
Query: 520 ---ALCKYASGTNPVTGVSTRVSLGCGNKRRNAYGTPSSPLLSAPPQKLLNRDGFCDEAT 350
ALCKYA+GTNP+TGV TR SLGCGN RRNAYG+PSSPLLSAPPQ+LL RDGFCDE T
Sbjct: 228 VLKALCKYATGTNPLTGVDTRKSLGCGNSRRNAYGSPSSPLLSAPPQRLLGRDGFCDEKT 287
Query: 349 GLCD 338
GLCD
Sbjct: 288 GLCD 291
>ref|NP_443033.1| unknown protein [Synechocystis sp. PCC 6803] gi|7469900|pir||S76933
hypothetical protein sll0577 - Synechocystis sp. (strain
PCC 6803) gi|1653935|dbj|BAA18845.1|
ORF_ID:sll0577~unknown protein [Synechocystis sp. PCC
6803]
Length = 169
Score = 63.9 bits (154), Expect = 2e-09
Identities = 32/57 (56%), Positives = 38/57 (66%)
Frame = -3
Query: 622 AILVRTVLTRSDLGGSIIEGADFSDAVLDLTQKLALCKYASGTNPVTGVSTRVSLGC 452
AIL ++ R+ + I+GADFS AVLD Q ALCK A G NP TG+STR SLGC
Sbjct: 112 AILAEAIMLRTSFKNAKIQGADFSLAVLDTEQIAALCKVADGVNPKTGISTRESLGC 168
>ref|ZP_00115441.1| hypothetical protein [Synechococcus sp. WH 8102]
Length = 190
Score = 61.2 bits (147), Expect = 1e-08
Identities = 31/57 (54%), Positives = 39/57 (68%)
Frame = -3
Query: 622 AILVRTVLTRSDLGGSIIEGADFSDAVLDLTQKLALCKYASGTNPVTGVSTRVSLGC 452
A +L +S + IEGADF+DAVLDL Q+ LC A+G +PV+GVSTR SLGC
Sbjct: 132 ATFTNAMLMQSRFADARIEGADFTDAVLDLPQQKLLCATAAGEHPVSGVSTRESLGC 188
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 507,832,620
Number of Sequences: 1393205
Number of extensions: 10502997
Number of successful extensions: 23774
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 23040
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23771
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)