Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005360A_C01 KMC005360A_c01
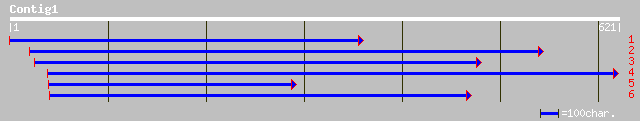
(581 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_176940.1| F12A21.13; protein id: At1g67740.1, supported b... 176 2e-43
gb|AAM65279.1| unknown [Arabidopsis thaliana] 176 2e-43
sp|P80470|PSBY_SPIOL Photosystem II core complex proteins psbY, ... 171 8e-42
ref|NP_060774.1| hypothetical protein FLJ11029 [Homo sapiens] gi... 42 0.006
gb|AAH08669.1|AAH08669 hypothetical protein FLJ11029 [Homo sapiens] 42 0.006
>ref|NP_176940.1| F12A21.13; protein id: At1g67740.1, supported by cDNA: 3813.,
supported by cDNA: gi_15215724, supported by cDNA:
gi_15529289, supported by cDNA: gi_20453250, supported
by cDNA: gi_3414927 [Arabidopsis thaliana]
gi|6093827|sp|O49347|PSBY_ARATH Photosystem II core
complex proteins psbY, chloroplast precursor (L-arginine
metabolising enzyme) (L-AME) [Contains: Photosystem II
protein psbY-1 (psbY-A1); Photosystem II protein psbY-2
(psbY-A2)] gi|11285513|pir||T51847 manganese-binding
protein PsbY precursor, photosystem II-associated
[imported] - Arabidopsis thaliana
gi|2956690|emb|CAA11248.1| PSBY [Arabidopsis thaliana]
gi|3414928|gb|AAC95011.1| PsbY precursor [Arabidopsis
thaliana] gi|11072011|gb|AAG28890.1|AC008113_6 F12A21.13
[Arabidopsis thaliana] gi|15215725|gb|AAK91408.1|
At1g67740/F12A21_13 [Arabidopsis thaliana]
gi|15529290|gb|AAK97739.1| At1g67740/F12A21_13
[Arabidopsis thaliana] gi|20453251|gb|AAM19864.1|
At1g67740/F12A21_13 [Arabidopsis thaliana]
gi|24417352|gb|AAN60286.1| unknown [Arabidopsis
thaliana]
Length = 189
Score = 176 bits (445), Expect = 2e-43
Identities = 97/157 (61%), Positives = 119/157 (75%), Gaps = 4/157 (2%)
Frame = -1
Query: 575 NTSVSTTSMAGTAIAGAIFSSLVSCDAAFAAQQIAEIA----EGDNRGLALLLPVIPAIA 408
N S++ TS TA+AGA+FSSL + A A QQIA++A DNRGLALLLP++PAIA
Sbjct: 40 NVSLAVTS---TALAGAVFSSLSYSEPALAIQQIAQLAAANASSDNRGLALLLPIVPAIA 96
Query: 407 WVLFNILQPALNQINRMRNSNGVIVGLGLGLGGLAAGSGMLWAPEASAGELGMIADAAAG 228
WVL+NILQPA+NQ+N+MR S G++VGLG+G GGLAA + PEA A A+AAA
Sbjct: 97 WVLYNILQPAINQVNKMRESKGIVVGLGIG-GGLAASGLLTPPPEAYAA-----AEAAAA 150
Query: 227 GSDNRGQLLLFVVAPAIGWVLFNILQPALNQINRMRS 117
SD+RGQLLL VV PA+ WVL+NILQPALNQIN+MRS
Sbjct: 151 SSDSRGQLLLIVVTPALLWVLYNILQPALNQINKMRS 187
Score = 60.5 bits (145), Expect = 2e-08
Identities = 34/71 (47%), Positives = 46/71 (63%), Gaps = 2/71 (2%)
Frame = -1
Query: 551 MAGTAIAGAIFSS--LVSCDAAFAAQQIAEIAEGDNRGLALLLPVIPAIAWVLFNILQPA 378
+ G I G + +S L A+AA + A A D+RG LL+ V PA+ WVL+NILQPA
Sbjct: 120 VVGLGIGGGLAASGLLTPPPEAYAAAEAAA-ASSDSRGQLLLIVVTPALLWVLYNILQPA 178
Query: 377 LNQINRMRNSN 345
LNQIN+MR+ +
Sbjct: 179 LNQINKMRSGD 189
>gb|AAM65279.1| unknown [Arabidopsis thaliana]
Length = 185
Score = 176 bits (445), Expect = 2e-43
Identities = 97/157 (61%), Positives = 119/157 (75%), Gaps = 4/157 (2%)
Frame = -1
Query: 575 NTSVSTTSMAGTAIAGAIFSSLVSCDAAFAAQQIAEIA----EGDNRGLALLLPVIPAIA 408
N S++ TS TA+AGA+FSSL + A A QQIA++A DNRGLALLLP++PAIA
Sbjct: 36 NVSLAVTS---TALAGAVFSSLSYSEPALAIQQIAQLAAANASSDNRGLALLLPIVPAIA 92
Query: 407 WVLFNILQPALNQINRMRNSNGVIVGLGLGLGGLAAGSGMLWAPEASAGELGMIADAAAG 228
WVL+NILQPA+NQ+N+MR S G++VGLG+G GGLAA + PEA A A+AAA
Sbjct: 93 WVLYNILQPAINQVNKMRESKGIVVGLGIG-GGLAASGLLTPPPEAYAA-----AEAAAA 146
Query: 227 GSDNRGQLLLFVVAPAIGWVLFNILQPALNQINRMRS 117
SD+RGQLLL VV PA+ WVL+NILQPALNQIN+MRS
Sbjct: 147 SSDSRGQLLLIVVTPALLWVLYNILQPALNQINKMRS 183
Score = 60.5 bits (145), Expect = 2e-08
Identities = 34/71 (47%), Positives = 46/71 (63%), Gaps = 2/71 (2%)
Frame = -1
Query: 551 MAGTAIAGAIFSS--LVSCDAAFAAQQIAEIAEGDNRGLALLLPVIPAIAWVLFNILQPA 378
+ G I G + +S L A+AA + A A D+RG LL+ V PA+ WVL+NILQPA
Sbjct: 116 VVGLGIGGGLAASGLLTPPPEAYAAAEAAA-ASSDSRGQLLLIVVTPALLWVLYNILQPA 174
Query: 377 LNQINRMRNSN 345
LNQIN+MR+ +
Sbjct: 175 LNQINKMRSGD 185
>sp|P80470|PSBY_SPIOL Photosystem II core complex proteins psbY, chloroplast precursor
(L-arginine metabolising enzyme) (L-AME) [Contains:
Photosystem II protein psbY-1 (psbY-A1); Photosystem II
protein psbY-2 (psbY-A2)] gi|7484673|pir||T08902
manganese-binding protein PsbY precursor, photosystem
II-associated - spinach gi|3337435|gb|AAC95000.1| PsbY
precursor; putative photosytem II peptide [Spinacia
oleracea]
Length = 199
Score = 171 bits (432), Expect = 8e-42
Identities = 93/146 (63%), Positives = 109/146 (73%), Gaps = 4/146 (2%)
Frame = -1
Query: 539 AIAGAIFSSLVSCDAAFAAQQIAEIAE----GDNRGLALLLPVIPAIAWVLFNILQPALN 372
AIAGA+F++L S D AFA QQ+A+IA DNRGLALLLP+IPA+ WVLFNILQPALN
Sbjct: 60 AIAGAVFATLGSVDPAFAVQQLADIAAEAGTSDNRGLALLLPIIPALGWVLFNILQPALN 119
Query: 371 QINRMRNSNGVIVGLGLGLGGLAAGSGMLWAPEASAGELGMIADAAAGGSDNRGQLLLFV 192
QIN+MRN + +GLGL GLA +L PEA A ++ A GSDNRG LLL V
Sbjct: 120 QINKMRNEKKAFI-VGLGLSGLATSGLLLATPEAQAA-----SEEIARGSDNRGTLLLLV 173
Query: 191 VAPAIGWVLFNILQPALNQINRMRSE 114
V PAIGWVLFNILQPALNQ+N+MRS+
Sbjct: 174 VLPAIGWVLFNILQPALNQLNKMRSQ 199
>ref|NP_060774.1| hypothetical protein FLJ11029 [Homo sapiens]
gi|7023440|dbj|BAA91964.1| unnamed protein product [Homo
sapiens]
Length = 360
Score = 42.0 bits (97), Expect = 0.006
Identities = 23/71 (32%), Positives = 35/71 (48%)
Frame = +1
Query: 145 AGCKMLNSTQPIAGATTNSRSCPLLSLPPAAASAIIPNSPADASGAHNIPEPAANPPSPS 324
+ C T ++G TN +C L++ P + A++P P A + P P PP P
Sbjct: 140 SSCPSCGQTCHMSGKLTNVPACVLIT--PGDSKAVLP--PTLPQPASHFPPPPPPPPLPP 195
Query: 325 PSPTITPLLLR 357
P P + P+LLR
Sbjct: 196 PPPPLAPVLLR 206
>gb|AAH08669.1|AAH08669 hypothetical protein FLJ11029 [Homo sapiens]
Length = 360
Score = 42.0 bits (97), Expect = 0.006
Identities = 23/71 (32%), Positives = 35/71 (48%)
Frame = +1
Query: 145 AGCKMLNSTQPIAGATTNSRSCPLLSLPPAAASAIIPNSPADASGAHNIPEPAANPPSPS 324
+ C T ++G TN +C L++ P + A++P P A + P P PP P
Sbjct: 140 SSCPSCGQTCHMSGKLTNVPACVLIT--PGDSKAVLP--PTLPQPASHFPPPPPPPPLPP 195
Query: 325 PSPTITPLLLR 357
P P + P+LLR
Sbjct: 196 PPPPLAPVLLR 206
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 565,136,339
Number of Sequences: 1393205
Number of extensions: 14254607
Number of successful extensions: 113405
Number of sequences better than 10.0: 562
Number of HSP's better than 10.0 without gapping: 71594
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 103175
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)