Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005345A_C01 KMC005345A_c01
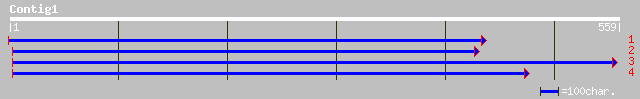
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_177755.1| hypothetical protein; protein id: At1g76280.1 [... 83 2e-15
gb|AAF17637.1|AC009978_13 T23E18.21 [Arabidopsis thaliana] 83 2e-15
dbj|BAA90802.1| Similar to putative salt-inducible protein (AC00... 34 1.1
ref|NP_464593.1| lmo1068 [Listeria monocytogenes EGD-e] gi|25381... 33 2.4
ref|NP_568460.1| putative protein; protein id: At5g24830.1 [Arab... 33 3.1
>ref|NP_177755.1| hypothetical protein; protein id: At1g76280.1 [Arabidopsis
thaliana]
Length = 812
Score = 83.2 bits (204), Expect = 2e-15
Identities = 42/88 (47%), Positives = 62/88 (69%)
Frame = -3
Query: 513 MQMQVVKALCSGERKKASDLLLDFGYRSRSLKADDFVGILKYCARSPDPLFVVEIWRLME 334
+Q+Q+V AL SGER+ AS LL + SL ADDF IL YCARSPDP+FV+E + +M
Sbjct: 57 LQLQIVDALRSGERQGASALLFKLIQGNYSLSADDFHDILYYCARSPDPVFVMETYSVMC 116
Query: 333 LKDVSMNNICSSLMMKALCKGGYLDEVN 250
K++S+++ ++K+LC GG+LD+ +
Sbjct: 117 KKEISLDSRSLLFIVKSLCNGGHLDKAS 144
>gb|AAF17637.1|AC009978_13 T23E18.21 [Arabidopsis thaliana]
Length = 800
Score = 83.2 bits (204), Expect = 2e-15
Identities = 42/88 (47%), Positives = 62/88 (69%)
Frame = -3
Query: 513 MQMQVVKALCSGERKKASDLLLDFGYRSRSLKADDFVGILKYCARSPDPLFVVEIWRLME 334
+Q+Q+V AL SGER+ AS LL + SL ADDF IL YCARSPDP+FV+E + +M
Sbjct: 16 LQLQIVDALRSGERQGASALLFKLIQGNYSLSADDFHDILYYCARSPDPVFVMETYSVMC 75
Query: 333 LKDVSMNNICSSLMMKALCKGGYLDEVN 250
K++S+++ ++K+LC GG+LD+ +
Sbjct: 76 KKEISLDSRSLLFIVKSLCNGGHLDKAS 103
>dbj|BAA90802.1| Similar to putative salt-inducible protein (AC006248) [Oryza sativa
(japonica cultivar-group)]
Length = 535
Score = 34.3 bits (77), Expect = 1.1
Identities = 28/98 (28%), Positives = 45/98 (45%), Gaps = 5/98 (5%)
Frame = -3
Query: 534 AGISPWSMQMQVVKALCSGERK--KASDLL---LDFGYRSRSLKADDFVGILKYCARSPD 370
A + P + VV C E K +A D + L GY + V +L +S +
Sbjct: 292 ARVQPNEVTYSVVIEACCKEEKPIEARDFMREMLGAGYVPDTALGAKVVDVLCQDGKSEE 351
Query: 369 PLFVVEIWRLMELKDVSMNNICSSLMMKALCKGGYLDE 256
++WR ME K+V +N+ +S ++ LCK G + E
Sbjct: 352 ---AYQLWRWMEKKNVPPDNMVTSTLIYWLCKNGMVRE 386
>ref|NP_464593.1| lmo1068 [Listeria monocytogenes EGD-e] gi|25381320|pir||AD1208
hypothetical protein lmo1068 [imported] - Listeria
monocytogenes (strain EGD-e) gi|16410470|emb|CAC99146.1|
lmo1068 [Listeria monocytogenes]
Length = 286
Score = 33.1 bits (74), Expect = 2.4
Identities = 23/72 (31%), Positives = 36/72 (49%)
Frame = +2
Query: 323 TSFNSINLQISTTNNGSGDRAQYFKIPTKSSAFNERLL*PKSKSKSEAFFLSPEQSALTT 502
T N N + +TT+ GS + + K T ++ + PKS +K+EAF SP ++T
Sbjct: 55 TKENDTNDKAATTDKGSSNPTKEEK-DTSTTTDTPKKETPKSPAKTEAFSFSPSGFKVST 113
Query: 503 CICIDHGDIPAT 538
I GD+ T
Sbjct: 114 VESILGGDVTTT 125
>ref|NP_568460.1| putative protein; protein id: At5g24830.1 [Arabidopsis thaliana]
gi|22655280|gb|AAM98230.1| putative protein [Arabidopsis
thaliana]
Length = 593
Score = 32.7 bits (73), Expect = 3.1
Identities = 18/83 (21%), Positives = 40/83 (47%), Gaps = 5/83 (6%)
Frame = -3
Query: 501 VVKALCS-----GERKKASDLLLDFGYRSRSLKADDFVGILKYCARSPDPLFVVEIWRLM 337
+V ALC KK + +LD + L ++ C ++ + + +E+W+ M
Sbjct: 232 IVHALCQKGVIGNNNKKLLEEILDSSQANAPLDIVICTILMDSCFKNGNVVQALEVWKEM 291
Query: 336 ELKDVSMNNICSSLMMKALCKGG 268
K+V +++ +++++ LC G
Sbjct: 292 SQKNVPADSVVYNVIIRGLCSSG 314
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 457,418,553
Number of Sequences: 1393205
Number of extensions: 9277060
Number of successful extensions: 20891
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 20314
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20890
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)