Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005329A_C01 KMC005329A_c01
(633 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
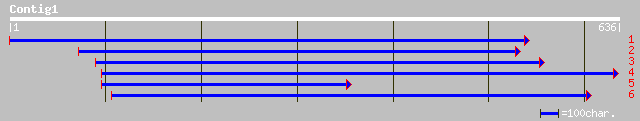
ref|NP_201522.1| putative protein; protein id: At5g67210.1 [Arab... 175 5e-43
gb|AAM64983.1| unknown [Arabidopsis thaliana] 172 3e-42
ref|NP_190591.1| putative protein; protein id: At3g50220.1, supp... 172 3e-42
ref|NP_565374.1| expressed protein; protein id: At2g15440.1, sup... 167 8e-41
emb|CAC09355.1| putative protein [Oryza sativa (indica cultivar-... 150 1e-35
>ref|NP_201522.1| putative protein; protein id: At5g67210.1 [Arabidopsis thaliana]
gi|10177608|dbj|BAB10955.1|
emb|CAB62301.1~gene_id:K21H1.17~similar to unknown
protein [Arabidopsis thaliana]
gi|27765046|gb|AAO23644.1| At5g67210 [Arabidopsis
thaliana]
Length = 317
Score = 175 bits (443), Expect = 5e-43
Identities = 85/146 (58%), Positives = 110/146 (75%), Gaps = 2/146 (1%)
Frame = -1
Query: 633 TKVSEHAQLLSQARAESTGDCRPVQNLLFSECKLGINDLPNHIYQISWDVILIDGPRGYS 454
TK E +L+S + + +CRPVQNLLFS+CKLG+NDLPNH+Y + WDVIL+DGPRG
Sbjct: 169 TKAREARELVSAVKEAARNECRPVQNLLFSDCKLGLNDLPNHVYDVDWDVILVDGPRGDG 228
Query: 453 AANPGRMSAIFTAAVLARSKKSGS--THVFVHDFKREVERIFSDEFLCQENLVQNVDQLG 280
PGRMS+IFTAAVLARSKK G+ THVFVHD+ R+VER+ DEFLC+ENLV++ D L
Sbjct: 229 GDVPGRMSSIFTAAVLARSKKGGNPKTHVFVHDYYRDVERLCGDEFLCRENLVESNDLLA 288
Query: 279 HFVVKSEKGGSSSQFCRNSSSLLSLS 202
H+V++ + +S+QFCR S+S
Sbjct: 289 HYVLE-KMDKNSTQFCRGRKKKRSVS 313
>gb|AAM64983.1| unknown [Arabidopsis thaliana]
Length = 322
Score = 172 bits (436), Expect = 3e-42
Identities = 87/149 (58%), Positives = 114/149 (76%), Gaps = 5/149 (3%)
Frame = -1
Query: 633 TKVSEHAQLLSQARAESTGDCRPVQNLLFSECKLGINDLPNHIYQISWDVILIDGPRGYS 454
TK E +L++ A+ + +CRPVQNLLFS+CKLG+NDLPNH+Y + WDVI +DGPRG +
Sbjct: 176 TKAHEAGELVTAAKEAAGNECRPVQNLLFSDCKLGLNDLPNHVYDVDWDVIFVDGPRGDA 235
Query: 453 AANPGRMSAIFTAAVLARSKKSGS--THVFVHDFKREVERIFSDEFLCQENLVQNVDQLG 280
PGRMS+IFTAAVLARSKK G+ THVFVHD+ R+VER+ DEFLC+ENLV++ D L
Sbjct: 236 HEGPGRMSSIFTAAVLARSKKGGNPKTHVFVHDYYRDVERLCGDEFLCRENLVESNDLLA 295
Query: 279 HFVV-KSEKGGSSSQFC--RNSSSLLSLS 202
H+V+ K +K +S++FC R S+ SLS
Sbjct: 296 HYVLDKMDK--NSTKFCNGRKKRSVSSLS 322
>ref|NP_190591.1| putative protein; protein id: At3g50220.1, supported by cDNA:
35440. [Arabidopsis thaliana] gi|11282604|pir||T45568
hypothetical protein F11C1.60 - Arabidopsis thaliana
gi|6523033|emb|CAB62301.1| putative protein [Arabidopsis
thaliana]
Length = 322
Score = 172 bits (436), Expect = 3e-42
Identities = 87/149 (58%), Positives = 114/149 (76%), Gaps = 5/149 (3%)
Frame = -1
Query: 633 TKVSEHAQLLSQARAESTGDCRPVQNLLFSECKLGINDLPNHIYQISWDVILIDGPRGYS 454
TK E +L++ A+ + +CRPVQNLLFS+CKLG+NDLPNH+Y + WDVI +DGPRG +
Sbjct: 176 TKAHEAGELVTAAKEAAGNECRPVQNLLFSDCKLGLNDLPNHVYDVDWDVIFVDGPRGDA 235
Query: 453 AANPGRMSAIFTAAVLARSKKSGS--THVFVHDFKREVERIFSDEFLCQENLVQNVDQLG 280
PGRMS+IFTAAVLARSKK G+ THVFVHD+ R+VER+ DEFLC+ENLV++ D L
Sbjct: 236 HEGPGRMSSIFTAAVLARSKKGGTPKTHVFVHDYYRDVERLCGDEFLCRENLVESNDLLA 295
Query: 279 HFVV-KSEKGGSSSQFC--RNSSSLLSLS 202
H+V+ K +K +S++FC R S+ SLS
Sbjct: 296 HYVLDKMDK--NSTKFCNGRKKRSVSSLS 322
>ref|NP_565374.1| expressed protein; protein id: At2g15440.1, supported by cDNA:
32729. [Arabidopsis thaliana] gi|25354650|pir||A84529
hypothetical protein At2g15440 [imported] - Arabidopsis
thaliana gi|4544369|gb|AAD22280.1| expressed protein
[Arabidopsis thaliana] gi|21592803|gb|AAM64752.1|
unknown [Arabidopsis thaliana]
Length = 329
Score = 167 bits (424), Expect = 8e-41
Identities = 84/146 (57%), Positives = 110/146 (74%), Gaps = 6/146 (4%)
Frame = -1
Query: 633 TKVSEHAQLLSQARAESTGDCRPVQNLLFSECKLGINDLPNHIYQISWDVILIDGPRGYS 454
TKVS+ +LL + +CRPVQNLLFS+CKLGINDLPN +Y+I WDVILIDGPRGY+
Sbjct: 166 TKVSQAKKLLGYYKTRP--ECRPVQNLLFSDCKLGINDLPNFVYEIDWDVILIDGPRGYA 223
Query: 453 AANPGRMSAIFTAAVLARSK----KSGSTHVFVHDFKREVERIFSDEFLCQENLVQNVDQ 286
+ +PGRM+ IFT+AVLA+SK K+ T V VH+F R++ER++S+EFLC+ENL++ V
Sbjct: 224 SDSPGRMAPIFTSAVLAKSKDFGTKTKKTDVLVHEFGRKIERVYSEEFLCEENLIEVVGD 283
Query: 285 LGHFVVKS--EKGGSSSQFCRNSSSL 214
LGHFVV + E+ FCRNS+ L
Sbjct: 284 LGHFVVAAAEERESYGDGFCRNSTRL 309
>emb|CAC09355.1| putative protein [Oryza sativa (indica cultivar-group)]
Length = 324
Score = 150 bits (379), Expect = 1e-35
Identities = 77/156 (49%), Positives = 100/156 (63%), Gaps = 5/156 (3%)
Frame = -1
Query: 633 TKVSEHAQLLSQARAESTGDCRPVQNLLFSECKLGINDLPNHIYQISWDVILIDGPRGYS 454
TKV + LL ARA +CRP+QNLLFSEC+L INDLPN +Y ++WD++LIDGP G++
Sbjct: 164 TKVRDFRDLLDAARASRAAECRPIQNLLFSECRLAINDLPNDLYDVAWDIVLIDGPSGWN 223
Query: 453 AANPGRMSAIFTAAVLAR---SKKSGSTHVFVHDFKREVERIFSDEFLCQENLV--QNVD 289
+PGRM +IFT AVLAR + G T V VHDF+ E+E++ S EFLC EN V
Sbjct: 224 PTSPGRMPSIFTTAVLARTGATAAKGPTDVLVHDFQFELEQVLSKEFLCDENRVAGSGTP 283
Query: 288 QLGHFVVKSEKGGSSSQFCRNSSSLLSLSVSKVSAK 181
LGHFVV+ + G FC S + + S+ S K
Sbjct: 284 SLGHFVVRPD--GRRDAFCSGQDSTAAGTSSEKSGK 317
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 546,281,751
Number of Sequences: 1393205
Number of extensions: 12374561
Number of successful extensions: 37925
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 35615
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37789
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)