Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005312A_C02 KMC005312A_c02
(945 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
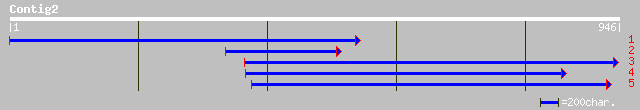
ref|NP_084777.1| ribosomal protein S12 [Lotus japonicus] gi|1351... 83 7e-15
sp|P07134|RR12_SOYBN Chloroplast 30S ribosomal protein S12 gi|32... 81 3e-14
ref|NP_051038.1| ribosomal protein S12 [Arabidopsis thaliana] gi... 79 8e-14
emb|CAA27361.1| rps12 (aa 1-38) [Nicotiana tabacum] gi|295336|gb... 79 8e-14
ref|NP_084768.1| ribosomal protein S12 [Oenothera elata subsp. h... 79 8e-14
>ref|NP_084777.1| ribosomal protein S12 [Lotus japonicus]
gi|13518483|ref|NP_084776.1| ribosomal protein S12
[Lotus japonicus] gi|18202718|sp|Q9B133|RR12_LOTJA
Chloroplast 30S ribosomal protein S12
gi|13359003|dbj|BAB33220.1| ribosomal protein S12 [Lotus
japonicus] gi|13359024|dbj|BAB33241.1| ribosomal protein
S12 [Lotus japonicus]
Length = 123
Score = 82.8 bits (203), Expect = 7e-15
Identities = 38/38 (100%), Positives = 38/38 (100%)
Frame = -3
Query: 778 MPTMKQLIRKTRQPIRNVTKSPALRGCPQRRGTCTRVY 665
MPTMKQLIRKTRQPIRNVTKSPALRGCPQRRGTCTRVY
Sbjct: 1 MPTMKQLIRKTRQPIRNVTKSPALRGCPQRRGTCTRVY 38
>sp|P07134|RR12_SOYBN Chloroplast 30S ribosomal protein S12 gi|320170|pir||A26574
ribosomal protein S12, chloroplast - soybean chloroplast
gi|11572|emb|CAA28661.1| rps12 [Glycine max]
Length = 123
Score = 80.9 bits (198), Expect = 3e-14
Identities = 37/38 (97%), Positives = 37/38 (97%)
Frame = -3
Query: 778 MPTMKQLIRKTRQPIRNVTKSPALRGCPQRRGTCTRVY 665
MPTMKQLIR TRQPIRNVTKSPALRGCPQRRGTCTRVY
Sbjct: 1 MPTMKQLIRNTRQPIRNVTKSPALRGCPQRRGTCTRVY 38
>ref|NP_051038.1| ribosomal protein S12 [Arabidopsis thaliana]
gi|7525080|ref|NP_051037.1| ribosomal protein S12
[Arabidopsis thaliana] gi|11497571|ref|NP_054910.1|
ribosomal protein S12 [Spinacia oleracea]
gi|11497594|ref|NP_054911.1| ribosomal protein S12
[Spinacia oleracea] gi|548847|sp|P06369|RR12_ARATH
Chloroplast 30S ribosomal protein S12
gi|1086182|pir||S39501 ribosomal protein S12 -
curled-leaved tobacco gi|5881718|dbj|BAA84409.1|
ribosomal protein S12 [Arabidopsis thaliana]
gi|5881741|dbj|BAA84432.1| ribosomal protein S12
[Arabidopsis thaliana] gi|7636152|emb|CAB88774.1|
ribosomal protein S12 [Spinacia oleracea]
gi|7636175|emb|CAB88797.1| ribosomal protein S12
[Spinacia oleracea] gi|225248|prf||1211235CG ribosomal
protein S12
Length = 123
Score = 79.3 bits (194), Expect = 8e-14
Identities = 36/38 (94%), Positives = 37/38 (96%)
Frame = -3
Query: 778 MPTMKQLIRKTRQPIRNVTKSPALRGCPQRRGTCTRVY 665
MPT+KQLIR TRQPIRNVTKSPALRGCPQRRGTCTRVY
Sbjct: 1 MPTIKQLIRNTRQPIRNVTKSPALRGCPQRRGTCTRVY 38
>emb|CAA27361.1| rps12 (aa 1-38) [Nicotiana tabacum] gi|295336|gb|AAA16345.1|
ribosomal protein S12 gi|5360577|dbj|BAA82064.1| 30S
ribosomal protein [Arabidopsis thaliana]
gi|13276711|emb|CAC34260.1| ribosomal protein S12
[Oenothera elata subsp. hookeri]
Length = 38
Score = 79.3 bits (194), Expect = 8e-14
Identities = 36/38 (94%), Positives = 37/38 (96%)
Frame = -3
Query: 778 MPTMKQLIRKTRQPIRNVTKSPALRGCPQRRGTCTRVY 665
MPT+KQLIR TRQPIRNVTKSPALRGCPQRRGTCTRVY
Sbjct: 1 MPTIKQLIRNTRQPIRNVTKSPALRGCPQRRGTCTRVY 38
>ref|NP_084768.1| ribosomal protein S12 [Oenothera elata subsp. hookeri]
gi|14423921|sp|Q9MDK3|RR12_OENHO Chloroplast 30S
ribosomal protein S12 gi|6723773|emb|CAB67182.1|
ribosomal protein S12 [Oenothera elata subsp. hookeri]
Length = 123
Score = 79.3 bits (194), Expect = 8e-14
Identities = 36/38 (94%), Positives = 37/38 (96%)
Frame = -3
Query: 778 MPTMKQLIRKTRQPIRNVTKSPALRGCPQRRGTCTRVY 665
MPT+KQLIR TRQPIRNVTKSPALRGCPQRRGTCTRVY
Sbjct: 1 MPTIKQLIRNTRQPIRNVTKSPALRGCPQRRGTCTRVY 38
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 748,680,851
Number of Sequences: 1393205
Number of extensions: 16212947
Number of successful extensions: 30087
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 28607
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29988
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 52969081112
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)