Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005304A_C01 KMC005304A_c01
(856 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
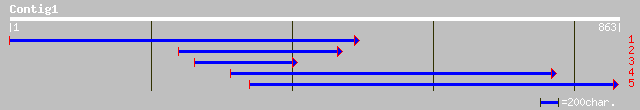
ref|NP_188606.1| hypothetical protein; protein id: At3g19720.1 [... 191 1e-47
pir||T52402 hypothetical protein MMB12.21 [imported] - Arabidops... 191 1e-47
dbj|BAC55068.1| dynamin related protein involved in chloroplast ... 80 3e-14
ref|NP_704789.1| hypothetical protein [Plasmodium falciparum 3D7... 37 0.37
gb|AAM88910.1| fast myosin heavy chain HCIII [Gallus gallus] 35 1.4
>ref|NP_188606.1| hypothetical protein; protein id: At3g19720.1 [Arabidopsis
thaliana]
Length = 649
Score = 191 bits (485), Expect = 1e-47
Identities = 93/117 (79%), Positives = 108/117 (91%)
Frame = -3
Query: 854 ETKLADLLDSTLWNRRLAPSSERIVYGLVQQIFHGIREYFLVSTELKFNCFLLMPIVDKL 675
E +LADLLDSTLWNR+LAPSSERIVY LVQQIF GIREYFL S ELKFNCFLLMPIVDKL
Sbjct: 517 EMRLADLLDSTLWNRKLAPSSERIVYALVQQIFQGIREYFLASAELKFNCFLLMPIVDKL 576
Query: 674 PARLREDLESAFEDDLDNVFDITNLQHSLGQQKRDTEIELKRIKRLREKFRMIHEQL 504
PA LRE+LE+AFEDDLD++FDITNL+ SL Q+KR TEIEL+RIKR++EKFR+++E+L
Sbjct: 577 PALLREELENAFEDDLDSIFDITNLRQSLDQKKRSTEIELRRIKRIKEKFRVMNEKL 633
>pir||T52402 hypothetical protein MMB12.21 [imported] - Arabidopsis thaliana
gi|9294439|dbj|BAB02559.1| gene_id:MMB12.21~unknown
protein [Arabidopsis thaliana]
Length = 772
Score = 191 bits (485), Expect = 1e-47
Identities = 93/117 (79%), Positives = 108/117 (91%)
Frame = -3
Query: 854 ETKLADLLDSTLWNRRLAPSSERIVYGLVQQIFHGIREYFLVSTELKFNCFLLMPIVDKL 675
E +LADLLDSTLWNR+LAPSSERIVY LVQQIF GIREYFL S ELKFNCFLLMPIVDKL
Sbjct: 640 EMRLADLLDSTLWNRKLAPSSERIVYALVQQIFQGIREYFLASAELKFNCFLLMPIVDKL 699
Query: 674 PARLREDLESAFEDDLDNVFDITNLQHSLGQQKRDTEIELKRIKRLREKFRMIHEQL 504
PA LRE+LE+AFEDDLD++FDITNL+ SL Q+KR TEIEL+RIKR++EKFR+++E+L
Sbjct: 700 PALLREELENAFEDDLDSIFDITNLRQSLDQKKRSTEIELRRIKRIKEKFRVMNEKL 756
>dbj|BAC55068.1| dynamin related protein involved in chloroplast division
[Cyanidioschyzon merolae]
Length = 962
Score = 80.5 bits (197), Expect = 3e-14
Identities = 41/114 (35%), Positives = 67/114 (57%)
Frame = -3
Query: 845 LADLLDSTLWNRRLAPSSERIVYGLVQQIFHGIREYFLVSTELKFNCFLLMPIVDKLPAR 666
+ +D L +R + PSS +V LV I R +F ELKFNCF LMP+V++ P
Sbjct: 786 ILQFMDDLLLSRVVTPSSMAVVSSLVDHIVDAWRMHFAKVVELKFNCFFLMPVVEEFPRF 845
Query: 665 LREDLESAFEDDLDNVFDITNLQHSLGQQKRDTEIELKRIKRLREKFRMIHEQL 504
RE L+ + DL++VF+I + +L +++ + E +R+++L+E F I+ QL
Sbjct: 846 CREKLDRLYGGDLNDVFNIAEARAALERRREELLQERRRVEKLQEMFNAINSQL 899
>ref|NP_704789.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23505151|emb|CAD51932.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 346
Score = 37.0 bits (84), Expect = 0.37
Identities = 23/73 (31%), Positives = 34/73 (46%)
Frame = +3
Query: 204 PCKRPTKNKDDTWSSI*NDLERSF*HSVFFHRTIPHASQSFSEINFQKLSSTNVVSLLHY 383
PC KN+D+ S N + S H+ F IPH Q I + L N+ L H
Sbjct: 190 PCNNYYKNQDNFLKS--NTVTYS--HNNLFKENIPHVHQHTGIITSKTLPQENIYVLKHV 245
Query: 384 SVPLFFGRKVHRD 422
S+P+F +++D
Sbjct: 246 SIPIFEKEIINKD 258
>gb|AAM88910.1| fast myosin heavy chain HCIII [Gallus gallus]
Length = 1940
Score = 35.0 bits (79), Expect = 1.4
Identities = 25/86 (29%), Positives = 41/86 (47%), Gaps = 3/86 (3%)
Frame = -3
Query: 749 IREYFLVSTELKFNCFLLMPIVDKLPARLREDLESAFE-DDLDNV--FDITNLQHSLGQQ 579
+RE S E + N L +VDKL +++ A E ++L NV +QH L +
Sbjct: 1851 VRELTYQSEEDRKNILRLQDLVDKLQMKVKSYKRQAEEAEELSNVNLSKFRKIQHELEEA 1910
Query: 578 KRDTEIELKRIKRLREKFRMIHEQLI 501
+ +I ++ +LR K R H + I
Sbjct: 1911 EERADIAESQVNKLRVKSREFHSKKI 1936
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 691,780,108
Number of Sequences: 1393205
Number of extensions: 14225547
Number of successful extensions: 31514
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 30248
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31470
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 45152354394
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)