Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005301A_C01 KMC005301A_c01
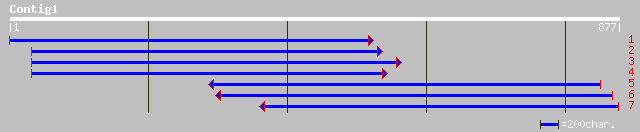
(868 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_198801.1| 40S ribosomal protein S9-like; protein id: At5g... 367 e-100
ref|NP_197024.1| 40S ribosomal protein - like; protein id: At5g1... 345 5e-94
dbj|BAA78592.1| 40S ribosomal protein S9 [Chlamydomonas sp. HS-5] 296 4e-79
sp|P52810|RS9_PODAN 40S ribosomal protein S9 (S7) gi|1321917|emb... 276 3e-73
gb|AAK95191.1|AF402817_1 40S ribosomal protein S9 [Ictalurus pun... 270 2e-71
>ref|NP_198801.1| 40S ribosomal protein S9-like; protein id: At5g39850.1, supported
by cDNA: 41408. [Arabidopsis thaliana]
gi|10176977|dbj|BAB10209.1| 40S ribosomal protein S9
[Arabidopsis thaliana] gi|21593688|gb|AAM65655.1| 40S
ribosomal protein S9-like [Arabidopsis thaliana]
Length = 197
Score = 367 bits (941), Expect = e-100
Identities = 180/197 (91%), Positives = 190/197 (96%)
Frame = -3
Query: 818 MVHVSFYRNYGKTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQYALSRIRNNARN 639
MV+V FYRNYGKTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQY LSRIRN AR
Sbjct: 1 MVNVRFYRNYGKTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQYTLSRIRNAARE 60
Query: 638 LLTLDEKNPRRIFEGEALLRRMFRYGLLDETQNKLDYVLALTVENFLERRLQTLVFKSGM 459
LLTLDEKNPRRIFEGEALLRRM RYGLLDETQNKLDYVLALTVENFLERRLQT+VFKSGM
Sbjct: 61 LLTLDEKNPRRIFEGEALLRRMNRYGLLDETQNKLDYVLALTVENFLERRLQTIVFKSGM 120
Query: 458 AKSIHHARVLIRQRHIRVGRQVVNIPSFMVRVDSQKHIDFSLTSPLGGGRPGRVKRKNQK 279
AKSIHHARVLIRQRHIRVGRQ+VNIPSFMVRV+SQKH+DFSLTSP GGGRPGRVKR+N++
Sbjct: 121 AKSIHHARVLIRQRHIRVGRQLVNIPSFMVRVESQKHVDFSLTSPFGGGRPGRVKRRNER 180
Query: 278 AAAKKASGGDGDEEDED 228
A AKKASGGDGDE+DE+
Sbjct: 181 AGAKKASGGDGDEDDEE 197
>ref|NP_197024.1| 40S ribosomal protein - like; protein id: At5g15200.1 [Arabidopsis
thaliana] gi|11276530|pir||T49955 40S ribosomal
protein-like - Arabidopsis thaliana
gi|7671489|emb|CAB89330.1| 40S ribosomal protein-like
[Arabidopsis thaliana]
Length = 198
Score = 345 bits (885), Expect = 5e-94
Identities = 171/198 (86%), Positives = 189/198 (95%), Gaps = 3/198 (1%)
Frame = -3
Query: 818 MVHVSFYRNYGKTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQYALSRIRNNARN 639
MVHV +YRNYGKTFK PRRPYEKERLD+ELKLVGEYGLR KRELWRVQY+LSRIRN AR+
Sbjct: 1 MVHVCYYRNYGKTFKGPRRPYEKERLDSELKLVGEYGLRNKRELWRVQYSLSRIRNAARD 60
Query: 638 LLTLDEKNPRRIFEGEALLRRMFRYGLLDETQNKLDYVLALTVENFLERRLQTLVFKSGM 459
LLTLDEK+PRRIFEGEALLRRM RYGLLDE+QNKLDYVLALTVENFLERRLQT+VFKSGM
Sbjct: 61 LLTLDEKSPRRIFEGEALLRRMNRYGLLDESQNKLDYVLALTVENFLERRLQTIVFKSGM 120
Query: 458 AKSIHHARVLIRQRHIRVGRQVVNIPSFMVRVDSQKHIDFSLTSPLGGGRPGRVKRKNQK 279
AKSIHH+RVLIRQRHIRVG+Q+VNIPSFMVR+DSQKHIDF+LTSP GGGRPGRVKR+N+K
Sbjct: 121 AKSIHHSRVLIRQRHIRVGKQLVNIPSFMVRLDSQKHIDFALTSPFGGGRPGRVKRRNEK 180
Query: 278 AAAKKASGG---DGDEED 234
+A+KKASGG DGD+E+
Sbjct: 181 SASKKASGGGDADGDDEE 198
>dbj|BAA78592.1| 40S ribosomal protein S9 [Chlamydomonas sp. HS-5]
Length = 192
Score = 296 bits (757), Expect = 4e-79
Identities = 147/196 (75%), Positives = 168/196 (85%)
Frame = -3
Query: 818 MVHVSFYRNYGKTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQYALSRIRNNARN 639
M + YRNYGKTFKKPRRP+EKERLD ELKLVGEYGLR KRELWRVQ LS+IRN ARN
Sbjct: 1 MPKIGQYRNYGKTFKKPRRPFEKERLDGELKLVGEYGLRNKRELWRVQMILSKIRNAARN 60
Query: 638 LLTLDEKNPRRIFEGEALLRRMFRYGLLDETQNKLDYVLALTVENFLERRLQTLVFKSGM 459
LL DE + +R+FEG+AL+RRM++YGLL+ETQ+KLDYVLAL +FLERRLQT+VFK G+
Sbjct: 61 LLQHDEDDTKRLFEGQALMRRMYKYGLLNETQDKLDYVLALQPNDFLERRLQTVVFKQGL 120
Query: 458 AKSIHHARVLIRQRHIRVGRQVVNIPSFMVRVDSQKHIDFSLTSPLGGGRPGRVKRKNQK 279
AKSIHHARVLI+Q HIRVG+QVVNIPSFMVRVDSQKH+DF+LTSP GGGRPGR KRKN
Sbjct: 121 AKSIHHARVLIKQNHIRVGKQVVNIPSFMVRVDSQKHVDFALTSPFGGGRPGRCKRKN-- 178
Query: 278 AAAKKASGGDGDEEDE 231
A K +GG G E+DE
Sbjct: 179 --AGKGAGGGGGEDDE 192
>sp|P52810|RS9_PODAN 40S ribosomal protein S9 (S7) gi|1321917|emb|CAA65433.1|
cytoplasmic ribosomal protein S7 [Podospora anserina]
Length = 190
Score = 276 bits (706), Expect = 3e-73
Identities = 139/191 (72%), Positives = 163/191 (84%), Gaps = 1/191 (0%)
Frame = -3
Query: 797 RNYGKTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQYALSRIRNNARNLLTLDEK 618
R+Y KT K PRRP+E RLD+ELKLVGEYGLR KRE+WRV LS+IR AR LLTLDEK
Sbjct: 4 RSYSKTAKVPRRPFEAARLDSELKLVGEYGLRNKREVWRVLLTLSKIRRAARELLTLDEK 63
Query: 617 NPRRIFEGEALLRRMFRYGLLDETQNKLDYVLALTVENFLERRLQTLVFKSGMAKSIHHA 438
+P+R+FEG AL+RR+ R G+LDE++ KLDYVLAL E+FLERRLQTLV+K G+AKSIHHA
Sbjct: 64 DPKRLFEGNALIRRLVRVGVLDESRMKLDYVLALKAEDFLERRLQTLVYKLGLAKSIHHA 123
Query: 437 RVLIRQRHIRVGRQVVNIPSFMVRVDSQKHIDFSLTSPLGGGRPGRVKRKNQKAAAKKAS 258
RVLIRQRHIRVG+Q+VN+PSF+VR+DSQKHIDF+LTSP GGGRPGRV+RK AK A
Sbjct: 124 RVLIRQRHIRVGKQIVNVPSFVVRLDSQKHIDFALTSPFGGGRPGRVRRKK----AKAAE 179
Query: 257 GGDGD-EEDED 228
GGDGD EEDE+
Sbjct: 180 GGDGDAEEDEE 190
>gb|AAK95191.1|AF402817_1 40S ribosomal protein S9 [Ictalurus punctatus]
Length = 194
Score = 270 bits (691), Expect = 2e-71
Identities = 134/184 (72%), Positives = 154/184 (82%)
Frame = -3
Query: 785 KTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQYALSRIRNNARNLLTLDEKNPRR 606
KT+ PRRP+EK RLD ELKL+GEYGLR KRE+WRV++ L++IR AR LLTLDEK+PRR
Sbjct: 11 KTYVTPRRPFEKSRLDQELKLIGEYGLRNKREVWRVKFTLAKIRKAARELLTLDEKDPRR 70
Query: 605 IFEGEALLRRMFRYGLLDETQNKLDYVLALTVENFLERRLQTLVFKSGMAKSIHHARVLI 426
+FEG ALLRR+ R G+LDE + KLDY+L L VE+FLERRLQT VFK G+AKSIHHARVLI
Sbjct: 71 LFEGNALLRRLVRIGVLDEGKMKLDYILGLKVEDFLERRLQTQVFKLGLAKSIHHARVLI 130
Query: 425 RQRHIRVGRQVVNIPSFMVRVDSQKHIDFSLTSPLGGGRPGRVKRKNQKAAAKKASGGDG 246
RQRHIRV +QVVNIPSF+VR+DSQKHIDFSL SP GGGRPGRVKRKN K GGD
Sbjct: 131 RQRHIRVRKQVVNIPSFVVRLDSQKHIDFSLRSPYGGGRPGRVKRKNAKKGQGGLGGGDD 190
Query: 245 DEED 234
+EED
Sbjct: 191 EEED 194
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 719,514,635
Number of Sequences: 1393205
Number of extensions: 15876344
Number of successful extensions: 70784
Number of sequences better than 10.0: 1142
Number of HSP's better than 10.0 without gapping: 64193
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 70345
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 46267227342
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)