Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005294A_C01 KMC005294A_c01
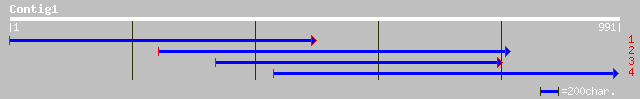
(991 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P35016|ENPL_CATRO Endoplasmin homolog precursor (GRP94 homolo... 551 e-156
dbj|BAB86368.1| SHEPHERD [Arabidopsis thaliana] 536 e-151
ref|NP_194150.1| HSP90-like protein; protein id: At4g24190.1, su... 536 e-151
gb|AAL79732.1|AC091774_23 heat shock protein 90 [Oryza sativa] 535 e-151
dbj|BAA90487.1| heat shock protein 90 [Oryza sativa] 532 e-150
>sp|P35016|ENPL_CATRO Endoplasmin homolog precursor (GRP94 homolog) gi|542022|pir||S39558
HSP90 homolog - Madagascar periwinkle
gi|348696|gb|AAA16785.1| heat shock protein 90
Length = 817
Score = 551 bits (1421), Expect = e-156
Identities = 279/326 (85%), Positives = 298/326 (90%), Gaps = 1/326 (0%)
Frame = +1
Query: 16 DFSDEKLYHGVTLLREGDVEFKAVLFVPPKAPQDSYESYYNSNKSNLKLFVRRVFISDEF 195
DFS+EK EGDVEFKA +PPKAPQD YESYYNSNKSNLKL+VRRVFISDEF
Sbjct: 374 DFSEEKPLAWSHFTAEGDVEFKAFTLLPPKAPQDLYESYYNSNKSNLKLYVRRVFISDEF 433
Query: 196 DEWLPKYLSFGKGLVDSDTLPLNVSREMLQAHSSLKTIKKKLIRKALDMIRRIADEDPDE 375
DE LPKYL+F KGLVDSDTLPLNVSREMLQ HSSLKTIKKKLIRKALDMIR+IADEDPDE
Sbjct: 434 DELLPKYLNFLKGLVDSDTLPLNVSREMLQQHSSLKTIKKKLIRKALDMIRKIADEDPDE 493
Query: 376 STDKEKKE-ETSSDIDEKRGQYAKFWNEFGKSIKLGIIEDATNRNR*AKLLRVETSKSEG 552
+ DK+KKE E S+D DEK+GQYAKFWNEFGKSIKLGIIEDA NRNR AKLLR E++KSEG
Sbjct: 494 ANDKDKKEVEESTDNDEKKGQYAKFWNEFGKSIKLGIIEDAANRNRLAKLLRFESTKSEG 553
Query: 553 KLTSLDQYISRMKAGQKDIFYITGSSKEQLENSPFLERLKKKNFEVIYFTDPVDEYLMQY 732
KLTSLDQYISRMK+GQKDIFYITG+SKEQLE SPFLERL KKN+EVI FTDPVDEYLMQY
Sbjct: 554 KLTSLDQYISRMKSGQKDIFYITGTSKEQLEKSPFLERLTKKNYEVILFTDPVDEYLMQY 613
Query: 733 LMDYEDKKFQNVSKEGLKLGKDTKDKELKESFKDLTKWWKNSLASENVDDVKISNRLDNT 912
LMDYEDKKFQNVSKEGLK+GKD+KDKELKESFK+LTKWWK +LASENVDDVKISNRL NT
Sbjct: 614 LMDYEDKKFQNVSKEGLKIGKDSKDKELKESFKELTKWWKGALASENVDDVKISNRLANT 673
Query: 913 PCVVVTSKYGWSANMERIMQSQTLSD 990
PCVVVTSKYGWS+NMERIMQSQTLSD
Sbjct: 674 PCVVVTSKYGWSSNMERIMQSQTLSD 699
>dbj|BAB86368.1| SHEPHERD [Arabidopsis thaliana]
Length = 823
Score = 536 bits (1382), Expect = e-151
Identities = 271/326 (83%), Positives = 293/326 (89%), Gaps = 1/326 (0%)
Frame = +1
Query: 16 DFSDEKLYHGVTLLREGDVEFKAVLFVPPKAPQDSYESYYNSNKSNLKLFVRRVFISDEF 195
DF+DEK EGDVEFKAVL+VPPKAP D YESYYNSNK+NLKL+VRRVFISDEF
Sbjct: 370 DFTDEKPMAWSHFNAEGDVEFKAVLYVPPKAPHDLYESYYNSNKANLKLYVRRVFISDEF 429
Query: 196 DEWLPKYLSFGKGLVDSDTLPLNVSREMLQAHSSLKTIKKKLIRKALDMIRRIADEDPDE 375
DE LPKYLSF KGLVDSDTLPLNVSREMLQ HSSLKTIKKKLIRKALDMIR++A+EDPDE
Sbjct: 430 DELLPKYLSFLKGLVDSDTLPLNVSREMLQQHSSLKTIKKKLIRKALDMIRKLAEEDPDE 489
Query: 376 STDKEKKE-ETSSDIDEKRGQYAKFWNEFGKSIKLGIIEDATNRNR*AKLLRVETSKSEG 552
D EKK+ E S + DEK+GQY KFWNEFGKS+KLGIIEDA NRNR AKLLR ET+KS+G
Sbjct: 490 IHDDEKKDVEKSGENDEKKGQYTKFWNEFGKSVKLGIIEDAANRNRLAKLLRFETTKSDG 549
Query: 553 KLTSLDQYISRMKAGQKDIFYITGSSKEQLENSPFLERLKKKNFEVIYFTDPVDEYLMQY 732
KLTSLDQYI RMK QKDIFYITGSSKEQLE SPFLERL KK +EVI+FTDPVDEYLMQY
Sbjct: 550 KLTSLDQYIKRMKKSQKDIFYITGSSKEQLEKSPFLERLIKKGYEVIFFTDPVDEYLMQY 609
Query: 733 LMDYEDKKFQNVSKEGLKLGKDTKDKELKESFKDLTKWWKNSLASENVDDVKISNRLDNT 912
LMDYEDKKFQNVSKEGLK+GKD+KDKELKE+FK+LTKWWK +LASENVDDVKISNRL +T
Sbjct: 610 LMDYEDKKFQNVSKEGLKVGKDSKDKELKEAFKELTKWWKGNLASENVDDVKISNRLADT 669
Query: 913 PCVVVTSKYGWSANMERIMQSQTLSD 990
PCVVVTSK+GWSANMERIMQSQTLSD
Sbjct: 670 PCVVVTSKFGWSANMERIMQSQTLSD 695
>ref|NP_194150.1| HSP90-like protein; protein id: At4g24190.1, supported by cDNA:
gi_14532541, supported by cDNA: gi_19570871 [Arabidopsis
thaliana] gi|7441896|pir||T09882 heat shock protein 90
homolog T22A6.20 - Arabidopsis thaliana
gi|5051761|emb|CAB45054.1| HSP90-like protein
[Arabidopsis thaliana] gi|7269269|emb|CAB79329.1|
HSP90-like protein [Arabidopsis thaliana]
gi|14532542|gb|AAK63999.1| AT4g24190/T22A6_20
[Arabidopsis thaliana] gi|19570872|dbj|BAB86369.1|
SHEPHERD [Arabidopsis thaliana]
gi|28416485|gb|AAO42773.1| At4g24190/T22A6_20
[Arabidopsis thaliana]
Length = 823
Score = 536 bits (1382), Expect = e-151
Identities = 271/326 (83%), Positives = 293/326 (89%), Gaps = 1/326 (0%)
Frame = +1
Query: 16 DFSDEKLYHGVTLLREGDVEFKAVLFVPPKAPQDSYESYYNSNKSNLKLFVRRVFISDEF 195
DF+DEK EGDVEFKAVL+VPPKAP D YESYYNSNK+NLKL+VRRVFISDEF
Sbjct: 370 DFTDEKPMAWSHFNAEGDVEFKAVLYVPPKAPHDLYESYYNSNKANLKLYVRRVFISDEF 429
Query: 196 DEWLPKYLSFGKGLVDSDTLPLNVSREMLQAHSSLKTIKKKLIRKALDMIRRIADEDPDE 375
DE LPKYLSF KGLVDSDTLPLNVSREMLQ HSSLKTIKKKLIRKALDMIR++A+EDPDE
Sbjct: 430 DELLPKYLSFLKGLVDSDTLPLNVSREMLQQHSSLKTIKKKLIRKALDMIRKLAEEDPDE 489
Query: 376 STDKEKKE-ETSSDIDEKRGQYAKFWNEFGKSIKLGIIEDATNRNR*AKLLRVETSKSEG 552
D EKK+ E S + DEK+GQY KFWNEFGKS+KLGIIEDA NRNR AKLLR ET+KS+G
Sbjct: 490 IHDDEKKDVEKSGENDEKKGQYTKFWNEFGKSVKLGIIEDAANRNRLAKLLRFETTKSDG 549
Query: 553 KLTSLDQYISRMKAGQKDIFYITGSSKEQLENSPFLERLKKKNFEVIYFTDPVDEYLMQY 732
KLTSLDQYI RMK QKDIFYITGSSKEQLE SPFLERL KK +EVI+FTDPVDEYLMQY
Sbjct: 550 KLTSLDQYIKRMKKSQKDIFYITGSSKEQLEKSPFLERLIKKGYEVIFFTDPVDEYLMQY 609
Query: 733 LMDYEDKKFQNVSKEGLKLGKDTKDKELKESFKDLTKWWKNSLASENVDDVKISNRLDNT 912
LMDYEDKKFQNVSKEGLK+GKD+KDKELKE+FK+LTKWWK +LASENVDDVKISNRL +T
Sbjct: 610 LMDYEDKKFQNVSKEGLKVGKDSKDKELKEAFKELTKWWKGNLASENVDDVKISNRLADT 669
Query: 913 PCVVVTSKYGWSANMERIMQSQTLSD 990
PCVVVTSK+GWSANMERIMQSQTLSD
Sbjct: 670 PCVVVTSKFGWSANMERIMQSQTLSD 695
>gb|AAL79732.1|AC091774_23 heat shock protein 90 [Oryza sativa]
Length = 812
Score = 535 bits (1379), Expect = e-151
Identities = 267/325 (82%), Positives = 293/325 (90%)
Frame = +1
Query: 16 DFSDEKLYHGVTLLREGDVEFKAVLFVPPKAPQDSYESYYNSNKSNLKLFVRRVFISDEF 195
DF D+K EGDVEFKA+LFVPPKAP D YESYYNSNKSNLKL+VRRVFISDEF
Sbjct: 372 DFGDDKPLSWSHFTAEGDVEFKALLFVPPKAPHDLYESYYNSNKSNLKLYVRRVFISDEF 431
Query: 196 DEWLPKYLSFGKGLVDSDTLPLNVSREMLQAHSSLKTIKKKLIRKALDMIRRIADEDPDE 375
DE LPKYLSF KGLVDSDTLPLNVSREMLQ HSSLKTIKKKLIRKALDMIR++A+EDPDE
Sbjct: 432 DELLPKYLSFLKGLVDSDTLPLNVSREMLQQHSSLKTIKKKLIRKALDMIRKLAEEDPDE 491
Query: 376 STDKEKKEETSSDIDEKRGQYAKFWNEFGKSIKLGIIEDATNRNR*AKLLRVETSKSEGK 555
++K+K +E S ++EK+GQYAKFWNEFGKS+KLGIIEDATNRNR AKLLR E++KSEGK
Sbjct: 492 YSNKDKTDEEKSAMEEKKGQYAKFWNEFGKSVKLGIIEDATNRNRLAKLLRFESTKSEGK 551
Query: 556 LTSLDQYISRMKAGQKDIFYITGSSKEQLENSPFLERLKKKNFEVIYFTDPVDEYLMQYL 735
L SLD+YISRMK GQKDIFYITGSSKEQLE SPFLERL KKN+EVIYFTDPVDEYLMQYL
Sbjct: 552 LASLDEYISRMKPGQKDIFYITGSSKEQLEKSPFLERLTKKNYEVIYFTDPVDEYLMQYL 611
Query: 736 MDYEDKKFQNVSKEGLKLGKDTKDKELKESFKDLTKWWKNSLASENVDDVKISNRLDNTP 915
MDYEDKKFQNVSKEGLKLGKD+K K+LKESFK+LT WWK +L +E+VD VKISNRL +TP
Sbjct: 612 MDYEDKKFQNVSKEGLKLGKDSKLKDLKESFKELTDWWKKALDTESVDSVKISNRLSDTP 671
Query: 916 CVVVTSKYGWSANMERIMQSQTLSD 990
CVVVTSKYGWSANME+IMQSQTLSD
Sbjct: 672 CVVVTSKYGWSANMEKIMQSQTLSD 696
>dbj|BAA90487.1| heat shock protein 90 [Oryza sativa]
Length = 810
Score = 532 bits (1371), Expect = e-150
Identities = 266/325 (81%), Positives = 292/325 (89%)
Frame = +1
Query: 16 DFSDEKLYHGVTLLREGDVEFKAVLFVPPKAPQDSYESYYNSNKSNLKLFVRRVFISDEF 195
DF D+K EGDVEFKA+LFVPPKAP D YESYYNSNKSNLKL+VRRV ISDEF
Sbjct: 370 DFGDDKPLSWSHFTAEGDVEFKALLFVPPKAPHDLYESYYNSNKSNLKLYVRRVSISDEF 429
Query: 196 DEWLPKYLSFGKGLVDSDTLPLNVSREMLQAHSSLKTIKKKLIRKALDMIRRIADEDPDE 375
DE LPKYLSF KGLVDSDTLPLNVSREMLQ HSSLKTIKKKLIRKALDMIR++A+EDPDE
Sbjct: 430 DELLPKYLSFLKGLVDSDTLPLNVSREMLQQHSSLKTIKKKLIRKALDMIRKLAEEDPDE 489
Query: 376 STDKEKKEETSSDIDEKRGQYAKFWNEFGKSIKLGIIEDATNRNR*AKLLRVETSKSEGK 555
++K+K +E S ++EK+GQYAKFWNEFGKS+KLGIIEDATNRNR AKLLR E++KSEGK
Sbjct: 490 YSNKDKTDEEKSAMEEKKGQYAKFWNEFGKSVKLGIIEDATNRNRLAKLLRFESTKSEGK 549
Query: 556 LTSLDQYISRMKAGQKDIFYITGSSKEQLENSPFLERLKKKNFEVIYFTDPVDEYLMQYL 735
L SLD+YISRMK GQKDIFYITGSSKEQLE SPFLERL KKN+EVIYFTDPVDEYLMQYL
Sbjct: 550 LASLDEYISRMKPGQKDIFYITGSSKEQLEKSPFLERLTKKNYEVIYFTDPVDEYLMQYL 609
Query: 736 MDYEDKKFQNVSKEGLKLGKDTKDKELKESFKDLTKWWKNSLASENVDDVKISNRLDNTP 915
MDYEDKKFQNVSKEGLKLGKD+K K+LKESFK+LT WWK +L +E+VD VKISNRL +TP
Sbjct: 610 MDYEDKKFQNVSKEGLKLGKDSKLKDLKESFKELTDWWKKALDTESVDSVKISNRLSDTP 669
Query: 916 CVVVTSKYGWSANMERIMQSQTLSD 990
CVVVTSKYGWSANME+IMQSQTLSD
Sbjct: 670 CVVVTSKYGWSANMEKIMQSQTLSD 694
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.314 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 813,401,420
Number of Sequences: 1393205
Number of extensions: 17396919
Number of successful extensions: 50664
Number of sequences better than 10.0: 538
Number of HSP's better than 10.0 without gapping: 45777
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49423
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 56566024535
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)