Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005239A_C01 KMC005239A_c01
(1056 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
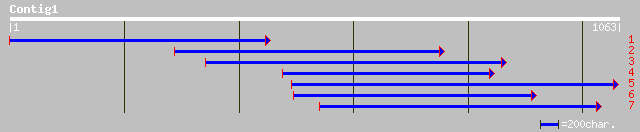
gb|AAF99798.1|AC012463_15 T2E6.19 [Arabidopsis thaliana] 178 1e-43
ref|NP_564513.1| expressed protein; protein id: At1g47740.1, sup... 178 1e-43
gb|AAM65611.1| unknown [Arabidopsis thaliana] 176 4e-43
ref|NP_568467.1| putative protein; protein id: At5g25170.1, supp... 102 1e-20
pir||E71444 probable EREBP-4 - Arabidopsis thaliana gi|2245108|e... 100 7e-20
>gb|AAF99798.1|AC012463_15 T2E6.19 [Arabidopsis thaliana]
Length = 292
Score = 178 bits (451), Expect = 1e-43
Identities = 83/132 (62%), Positives = 102/132 (76%), Gaps = 2/132 (1%)
Frame = -2
Query: 1055 REFMERLSADYYGDSYHLIVKNCNHFCDDICYKLTGNSIPKWVNRLARMGSYCHCVLPDA 876
REFME ++ YYG+ YHLIVKNCNHFC D+CYKLTG IPKWVNRLA++GS C C+LP++
Sbjct: 159 REFMEDMACSYYGNMYHLIVKNCNHFCQDVCYKLTGKKIPKWVNRLAQIGSVCSCILPES 218
Query: 875 LTTTTVQNDPDFQ--GCDSEKRRLRTAFSCLSSISMPHKEVSMSSLFLHSHYKGCLPPWE 702
L T V +DPD Q ++EKR LR++FSCLSSISM K++S SSLFL S +GCLPPW+
Sbjct: 219 LKITAVCHDPDGQIPEEENEKRSLRSSFSCLSSISMRQKQLSTSSLFLQSPLRGCLPPWQ 278
Query: 701 LKKSKKGSLKQK 666
LK+SK S K
Sbjct: 279 LKRSKSNSSSLK 290
>ref|NP_564513.1| expressed protein; protein id: At1g47740.1, supported by cDNA: 40816.
[Arabidopsis thaliana] gi|19424079|gb|AAL87252.1| unknown
protein [Arabidopsis thaliana] gi|21280795|gb|AAM45073.1|
unknown protein [Arabidopsis thaliana]
Length = 279
Score = 178 bits (451), Expect = 1e-43
Identities = 83/132 (62%), Positives = 102/132 (76%), Gaps = 2/132 (1%)
Frame = -2
Query: 1055 REFMERLSADYYGDSYHLIVKNCNHFCDDICYKLTGNSIPKWVNRLARMGSYCHCVLPDA 876
REFME ++ YYG+ YHLIVKNCNHFC D+CYKLTG IPKWVNRLA++GS C C+LP++
Sbjct: 146 REFMEDMACSYYGNMYHLIVKNCNHFCQDVCYKLTGKKIPKWVNRLAQIGSVCSCILPES 205
Query: 875 LTTTTVQNDPDFQ--GCDSEKRRLRTAFSCLSSISMPHKEVSMSSLFLHSHYKGCLPPWE 702
L T V +DPD Q ++EKR LR++FSCLSSISM K++S SSLFL S +GCLPPW+
Sbjct: 206 LKITAVCHDPDGQIPEEENEKRSLRSSFSCLSSISMRQKQLSTSSLFLQSPLRGCLPPWQ 265
Query: 701 LKKSKKGSLKQK 666
LK+SK S K
Sbjct: 266 LKRSKSNSSSLK 277
>gb|AAM65611.1| unknown [Arabidopsis thaliana]
Length = 251
Score = 176 bits (447), Expect = 4e-43
Identities = 82/132 (62%), Positives = 102/132 (77%), Gaps = 2/132 (1%)
Frame = -2
Query: 1055 REFMERLSADYYGDSYHLIVKNCNHFCDDICYKLTGNSIPKWVNRLARMGSYCHCVLPDA 876
REFME ++ YYG+ YHLIVKNCNHFC D+CYKLTG IPKWVNRLA++GS C C+LP++
Sbjct: 118 REFMEDMACSYYGNMYHLIVKNCNHFCQDVCYKLTGKKIPKWVNRLAQIGSVCSCILPES 177
Query: 875 LTTTTVQNDPDFQ--GCDSEKRRLRTAFSCLSSISMPHKEVSMSSLFLHSHYKGCLPPWE 702
L T V +DPD Q ++EKR LR++FSCLSSIS+ K++S SSLFL S +GCLPPW+
Sbjct: 178 LKITAVCHDPDGQIPEEENEKRSLRSSFSCLSSISIRQKQLSTSSLFLQSPLRGCLPPWQ 237
Query: 701 LKKSKKGSLKQK 666
LK+SK S K
Sbjct: 238 LKRSKSNSSSLK 249
>ref|NP_568467.1| putative protein; protein id: At5g25170.1, supported by cDNA: 263500.
[Arabidopsis thaliana]
Length = 218
Score = 102 bits (254), Expect = 1e-20
Identities = 56/122 (45%), Positives = 74/122 (59%), Gaps = 6/122 (4%)
Frame = -2
Query: 1055 REFMERLSADYYGDSYHLIVKNCNHFCDDICYKLTGNSIPKWVNRLARMGSYCHCVLPDA 876
R FME+L+ +Y G+SYHLI KNCNHFC+D+C +LT SIP WVNRLAR G +C+CVLP
Sbjct: 95 RVFMEKLAEEYSGNSYHLITKNCNHFCNDVCVQLTRRSIPSWVNRLARFGLFCNCVLPAE 154
Query: 875 LTTTTVQ--NDPDFQGCDSEKRRLRTAFSCLSSISMPHKEVSMSSLFLHS----HYKGCL 714
L T V+ + + + EK++LR+ SS P +S S S + CL
Sbjct: 155 LNETKVRQVRSKEEKIPEVEKKKLRSR----SSRFPPGPSLSSSGSLNRSRRGERRRQCL 210
Query: 713 PP 708
PP
Sbjct: 211 PP 212
>pir||E71444 probable EREBP-4 - Arabidopsis thaliana gi|2245108|emb|CAB10530.1|
EREBP-4 like protein [Arabidopsis thaliana]
gi|7268501|emb|CAB78752.1| EREBP-4 like protein
[Arabidopsis thaliana]
Length = 603
Score = 99.8 bits (247), Expect = 7e-20
Identities = 39/71 (54%), Positives = 56/71 (77%)
Frame = -2
Query: 1055 REFMERLSADYYGDSYHLIVKNCNHFCDDICYKLTGNSIPKWVNRLARMGSYCHCVLPDA 876
R +ME+LS Y+GD+YHLI KNCNHF +++C +LTG IP W+NRLAR+GS+C+C+LP++
Sbjct: 482 RSYMEKLSRKYHGDTYHLIAKNCNHFTEEVCLQLTGKPIPGWINRLARVGSFCNCLLPES 541
Query: 875 LTTTTVQNDPD 843
+ T V P+
Sbjct: 542 IQLTAVSALPE 552
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 887,253,094
Number of Sequences: 1393205
Number of extensions: 19513987
Number of successful extensions: 52242
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 48684
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 52122
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 62636524729
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)