Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005227A_C01 KMC005227A_c01
(708 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
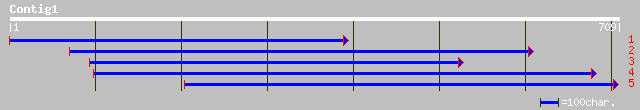
gb|AAD04752.1| phenylalkylamine binding protein homolog [Arabido... 160 1e-38
ref|NP_173433.1| C-8,7 sterol isomerase; protein id: At1g20050.1... 160 2e-38
gb|AAM63292.1| C-8,7 sterol isomerase [Arabidopsis thaliana] 160 2e-38
sp|Q9FTZ2|EBP_ORYSA Probable 3-beta-hydroxysteroid-delta(8),delt... 153 3e-36
gb|EAA28581.1| hypothetical protein [Neurospora crassa] 70 3e-11
>gb|AAD04752.1| phenylalkylamine binding protein homolog [Arabidopsis thaliana]
Length = 223
Score = 160 bits (406), Expect = 1e-38
Identities = 76/115 (66%), Positives = 93/115 (80%)
Frame = -3
Query: 706 YVGRDAGVVTVEGLTAVIEGPASLLAVYAIATGKSYSYMLQFAISLGQLYGTAVYYITAI 527
YVGRD+ VV+VEG+TAVI GPASLLA+YAIA KSYSY+LQ AIS+ QLYG VY+ITAI
Sbjct: 105 YVGRDSAVVSVEGITAVIVGPASLLAIYAIAKEKSYSYVLQLAISVCQLYGCVVYFITAI 164
Query: 526 LEGDNFSTNSFYYYAYYIGANFSWIVIPSIIAIRCWRKTSAAFRVQGQTKKPKVR 362
LEGDNF+TNSFYYY+YYIGAN W++IPS+I+ RCW+K AA + + K +
Sbjct: 165 LEGDNFATNSFYYYSYYIGANCWWVLIPSLISFRCWKKICAAAAIANNNVETKTK 219
>ref|NP_173433.1| C-8,7 sterol isomerase; protein id: At1g20050.1, supported by cDNA:
2121., supported by cDNA: gi_12248038, supported by
cDNA: gi_2772933, supported by cDNA: gi_4098988
[Arabidopsis thaliana] gi|18202046|sp|O48962|EBP_ARATH
Probable
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase
(Cholestenol delta-isomerase) (Delta8-delta7 sterol
isomerase) (D8-D7 sterol isomerase)
gi|11279073|pir||T51727 C-8,7 sterol isomerase
[validated] - Arabidopsis thaliana
gi|2772934|gb|AAD03489.1| C-8,7 sterol isomerase; aSI1
[Arabidopsis thaliana]
gi|8778994|gb|AAF79909.1|AC022472_18 Identical to C-8,7
sterol isomerase from Arabidopsis thaliana gb|AF030357.
ESTs gb|AI998831, gb|AA585846, gb|T22967 come from this
gene gi|12248039|gb|AAG50111.1|AF334733_1 putative C-8,7
sterol isomerase [Arabidopsis thaliana]
Length = 223
Score = 160 bits (405), Expect = 2e-38
Identities = 76/115 (66%), Positives = 93/115 (80%)
Frame = -3
Query: 706 YVGRDAGVVTVEGLTAVIEGPASLLAVYAIATGKSYSYMLQFAISLGQLYGTAVYYITAI 527
YVGRD+ VV+VEG+TAVI GPASLLA+YAIA KSYSY+LQ AIS+ QLYG VY+ITAI
Sbjct: 105 YVGRDSAVVSVEGITAVIVGPASLLAIYAIAKEKSYSYVLQLAISVCQLYGCLVYFITAI 164
Query: 526 LEGDNFSTNSFYYYAYYIGANFSWIVIPSIIAIRCWRKTSAAFRVQGQTKKPKVR 362
LEGDNF+TNSFYYY+YYIGAN W++IPS+I+ RCW+K AA + + K +
Sbjct: 165 LEGDNFATNSFYYYSYYIGANCWWVLIPSLISFRCWKKICAAAAIANNNVETKTK 219
>gb|AAM63292.1| C-8,7 sterol isomerase [Arabidopsis thaliana]
Length = 223
Score = 160 bits (405), Expect = 2e-38
Identities = 76/115 (66%), Positives = 93/115 (80%)
Frame = -3
Query: 706 YVGRDAGVVTVEGLTAVIEGPASLLAVYAIATGKSYSYMLQFAISLGQLYGTAVYYITAI 527
YVGRD+ VV+VEG+TAVI GPASLLA+YAIA KSYSY+LQ AIS+ QLYG VY+ITAI
Sbjct: 105 YVGRDSAVVSVEGITAVIVGPASLLAIYAIAKEKSYSYVLQLAISVCQLYGCLVYFITAI 164
Query: 526 LEGDNFSTNSFYYYAYYIGANFSWIVIPSIIAIRCWRKTSAAFRVQGQTKKPKVR 362
LEGDNF+TNSFYYY+YYIGAN W++IPS+I+ RCW+K AA + + K +
Sbjct: 165 LEGDNFATNSFYYYSYYIGANCWWVLIPSLISFRCWKKICAAAAIANNNVETKTK 219
>sp|Q9FTZ2|EBP_ORYSA Probable 3-beta-hydroxysteroid-delta(8),delta(7)-isomerase
(Cholestenol delta-isomerase) (Delta8-delta7 sterol
isomerase) (D8-D7 sterol isomerase)
gi|10697192|dbj|BAB16323.1| putative C-8,7 sterol
isomerase [Oryza sativa (japonica cultivar-group)]
gi|20804451|dbj|BAB92148.1| putative C-8,7 sterol
isomerase [Oryza sativa (japonica cultivar-group)]
Length = 219
Score = 153 bits (386), Expect = 3e-36
Identities = 75/113 (66%), Positives = 92/113 (81%)
Frame = -3
Query: 706 YVGRDAGVVTVEGLTAVIEGPASLLAVYAIATGKSYSYMLQFAISLGQLYGTAVYYITAI 527
YV RD VTVEG+TAV+EGPASLLAVYAIA+GKSYS++LQF + LGQLYG VY+ITA
Sbjct: 109 YVARDPATVTVEGITAVLEGPASLLAVYAIASGKSYSHILQFTVCLGQLYGCLVYFITAY 168
Query: 526 LEGDNFSTNSFYYYAYYIGANFSWIVIPSIIAIRCWRKTSAAFRVQGQTKKPK 368
L+G NF T+ FY++AY+IGAN SW+VIP++IAIR W+K AAF QG+ K K
Sbjct: 169 LDGFNFWTSPFYFWAYFIGANSSWVVIPTMIAIRSWKKICAAF--QGEKVKTK 219
>gb|EAA28581.1| hypothetical protein [Neurospora crassa]
Length = 345
Score = 70.1 bits (170), Expect = 3e-11
Identities = 39/119 (32%), Positives = 64/119 (53%), Gaps = 6/119 (5%)
Frame = -3
Query: 706 YVGRDAGVVTVEGLTAVIEGPASLLAVYAIATGK-SYSYMLQFAISLGQLYGTAVYYITA 530
Y+ D ++ +E LT + GP S LAV+AI G S ++ Q +S+G LYG A+Y+ T
Sbjct: 114 YMTSDPFMLCIETLTVLTWGPLSFLAVFAIIKGNTSLRHITQTIVSVGHLYGVALYFGTC 173
Query: 529 ILE----GDNFST-NSFYYYAYYIGANFSWIVIPSIIAIRCWRKTSAAFRVQGQTKKPK 368
+ G ++S + YY+ YY G N W+++P+I+ + + S V + K K
Sbjct: 174 FFQEKFRGISYSRPETLYYWVYYAGMNAPWVIVPAILLFQSSKTISQGLSVLNKVKSMK 232
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 565,319,163
Number of Sequences: 1393205
Number of extensions: 11775814
Number of successful extensions: 27157
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 26038
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27115
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32373034405
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)