Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005223A_C01 KMC005223A_c01
(858 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
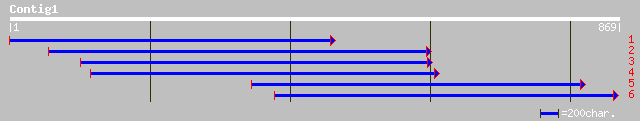
ref|NP_567334.1| inositol 1,3,4-trisphosphate 5/6-kinase-like pr... 145 3e-47
gb|AAO00682.1| Putative inositol 1,3,4-trisphosphate 5/6-kinase ... 133 5e-39
gb|AAK00417.2|AC069324_3 Putative inositol 1,3,4-trisphosphate 5... 133 6e-38
pir||T10544 inositol 1,3,4-trisphosphate 5/6-kinase homolog T12G... 96 2e-32
gb|AAG17052.1| saline-responsive OSSR1 [Oryza sativa] 106 6e-31
>ref|NP_567334.1| inositol 1,3,4-trisphosphate 5/6-kinase-like protein; protein id:
At4g08170.1, supported by cDNA: gi_15215757, supported
by cDNA: gi_16323319 [Arabidopsis thaliana]
gi|15215758|gb|AAK91424.1| AT4g08170/T12G13_10
[Arabidopsis thaliana] gi|16323320|gb|AAL15415.1|
AT4g08170/T12G13_10 [Arabidopsis thaliana]
Length = 353
Score = 145 bits (366), Expect(2) = 3e-47
Identities = 68/85 (80%), Positives = 78/85 (91%)
Frame = -2
Query: 734 ADLDPSLAVHPPRPLLEKLARELRHRLGLRLFNIDMIREYGTRDVFYVIDINYFPGYGKM 555
ADLDPS+A PPRPLLE+LA+ELR LGLRLFN+D+IRE+GTRD FYVIDINYFPGYGKM
Sbjct: 266 ADLDPSIAELPPRPLLERLAKELRRGLGLRLFNLDIIREHGTRDRFYVIDINYFPGYGKM 325
Query: 554 PEYEHVFTDFLLSLMQSKCKKKLTA 480
PEYEHVFTDFLLS++QS+CKK+ A
Sbjct: 326 PEYEHVFTDFLLSVVQSQCKKRALA 350
Score = 66.2 bits (160), Expect(2) = 3e-47
Identities = 33/41 (80%), Positives = 37/41 (89%)
Frame = -1
Query: 858 GEAIKVVRRFSLPILSKRELSKVAGLFRFP*VSCAAASADD 736
GEAI+VVRRFSLP +S+REL K AG+FRFP VSCAAASADD
Sbjct: 225 GEAIRVVRRFSLPDVSRRELPKSAGVFRFPRVSCAAASADD 265
>gb|AAO00682.1| Putative inositol 1,3,4-trisphosphate 5/6-kinase [Oryza sativa
(japonica cultivar-group)] gi|27356669|gb|AAO06958.1|
Putative inositol 1,3,4-trisphosphate 5/6-kinase [Oryza
sativa (japonica cultivar-group)]
Length = 357
Score = 133 bits (335), Expect(2) = 5e-39
Identities = 63/84 (75%), Positives = 73/84 (86%)
Frame = -2
Query: 734 ADLDPSLAVHPPRPLLEKLARELRHRLGLRLFNIDMIREYGTRDVFYVIDINYFPGYGKM 555
ADLDP ++ PPRPLLEKL +ELR RLGLRLFNIDMIRE GT+D +Y+IDINYFPG+GKM
Sbjct: 272 ADLDPHISELPPRPLLEKLGKELRGRLGLRLFNIDMIRELGTKDRYYIIDINYFPGFGKM 331
Query: 554 PEYEHVFTDFLLSLMQSKCKKKLT 483
P YEH+FTDFLL+L QSK KK L+
Sbjct: 332 PGYEHIFTDFLLNLAQSKYKKCLS 355
Score = 50.4 bits (119), Expect(2) = 5e-39
Identities = 25/40 (62%), Positives = 31/40 (77%)
Frame = -1
Query: 858 GEAIKVVRRFSLPILSKRELSKVAGLFRFP*VSCAAASAD 739
GE I+VVRRFSLP ++ +L G++RFP VSCAAASAD
Sbjct: 231 GETIQVVRRFSLPDVNTYDLLNNVGVYRFPRVSCAAASAD 270
>gb|AAK00417.2|AC069324_3 Putative inositol 1,3,4-trisphosphate 5/6-kinase [Oryza sativa]
Length = 333
Score = 133 bits (334), Expect(2) = 6e-38
Identities = 61/84 (72%), Positives = 74/84 (87%)
Frame = -2
Query: 734 ADLDPSLAVHPPRPLLEKLARELRHRLGLRLFNIDMIREYGTRDVFYVIDINYFPGYGKM 555
A++DPS+A PP+PLLEKL RELR RLGLRLFN DMIRE+G +D +YVIDINYFPGYGKM
Sbjct: 248 AEVDPSIAELPPKPLLEKLGRELRRRLGLRLFNFDMIREHGRKDRYYVIDINYFPGYGKM 307
Query: 554 PEYEHVFTDFLLSLMQSKCKKKLT 483
P YEH+F DFLLSL+Q+K K++L+
Sbjct: 308 PGYEHIFIDFLLSLVQNKYKRRLS 331
Score = 47.4 bits (111), Expect(2) = 6e-38
Identities = 23/41 (56%), Positives = 30/41 (73%)
Frame = -1
Query: 858 GEAIKVVRRFSLPILSKRELSKVAGLFRFP*VSCAAASADD 736
GE I+VVRRFSLP ++ +L G+FRFP VSCA +A+D
Sbjct: 207 GETIRVVRRFSLPDVNIYDLENNDGIFRFPRVSCATNTAED 247
>pir||T10544 inositol 1,3,4-trisphosphate 5/6-kinase homolog T12G13.10 -
Arabidopsis thaliana gi|5262190|emb|CAB45787.1| inositol
1, 3, 4-trisphosphate 5/6-kinase-like protein
[Arabidopsis thaliana] gi|7267457|emb|CAB81153.1|
inositol 1, 3, 4-trisphosphate 5/6-kinase-like protein
[Arabidopsis thaliana]
Length = 338
Score = 95.9 bits (237), Expect(2) = 2e-32
Identities = 45/56 (80%), Positives = 51/56 (90%)
Frame = -2
Query: 734 ADLDPSLAVHPPRPLLEKLARELRHRLGLRLFNIDMIREYGTRDVFYVIDINYFPG 567
ADLDPS+A PPRPLLE+LA+ELR LGLRLFN+D+IRE+GTRD FYVIDINYFPG
Sbjct: 266 ADLDPSIAELPPRPLLERLAKELRRGLGLRLFNLDIIREHGTRDRFYVIDINYFPG 321
Score = 66.2 bits (160), Expect(2) = 2e-32
Identities = 33/41 (80%), Positives = 37/41 (89%)
Frame = -1
Query: 858 GEAIKVVRRFSLPILSKRELSKVAGLFRFP*VSCAAASADD 736
GEAI+VVRRFSLP +S+REL K AG+FRFP VSCAAASADD
Sbjct: 225 GEAIRVVRRFSLPDVSRRELPKSAGVFRFPRVSCAAASADD 265
>gb|AAG17052.1| saline-responsive OSSR1 [Oryza sativa]
Length = 117
Score = 106 bits (265), Expect(2) = 6e-31
Identities = 49/64 (76%), Positives = 56/64 (86%)
Frame = -2
Query: 734 ADLDPSLAVHPPRPLLEKLARELRHRLGLRLFNIDMIREYGTRDVFYVIDINYFPGYGKM 555
ADLDP ++ PPRPLLEKL +ELR RLGLRLFNIDMIRE GT+D +Y+IDINYFPG+GKM
Sbjct: 47 ADLDPHISELPPRPLLEKLGKELRGRLGLRLFNIDMIRELGTKDRYYIIDINYFPGFGKM 106
Query: 554 PEYE 543
P YE
Sbjct: 107 PGYE 110
Score = 50.4 bits (119), Expect(2) = 6e-31
Identities = 25/40 (62%), Positives = 31/40 (77%)
Frame = -1
Query: 858 GEAIKVVRRFSLPILSKRELSKVAGLFRFP*VSCAAASAD 739
GE I+VVRRFSLP ++ +L G++RFP VSCAAASAD
Sbjct: 6 GETIQVVRRFSLPDVNTYDLLNNVGVYRFPRVSCAAASAD 45
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 724,477,258
Number of Sequences: 1393205
Number of extensions: 15676317
Number of successful extensions: 39097
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 37230
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39065
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 45431072631
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)