Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
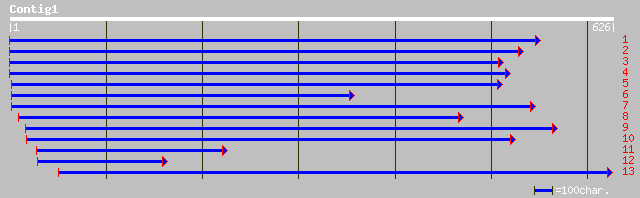
Query= KMC005221A_C01 KMC005221A_c01
(623 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181812.1| En/Spm-like transposon protein; protein id: At2... 193 1e-48
dbj|BAB90075.1| VsaA -like protein [Oryza sativa (japonica culti... 157 1e-37
dbj|BAB32984.1| P0489A05.4 [Oryza sativa (japonica cultivar-grou... 110 2e-23
ref|NP_115763.1| protein kinase, lysine deficient 4; putative pr... 45 0.001
dbj|BAC04669.1| unnamed protein product [Homo sapiens] 45 0.001
>ref|NP_181812.1| En/Spm-like transposon protein; protein id: At2g42840.1, supported
by cDNA: 41214. [Arabidopsis thaliana]
gi|25456394|pir||T52305 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana
gi|4512671|gb|AAD21725.1| En/Spm-like transposon protein
[Arabidopsis thaliana]
gi|4929128|gb|AAD33868.1|AF141375_1 protodermal factor 1
[Arabidopsis thaliana]
gi|4929130|gb|AAD33869.1|AF141376_1 protodermal factor 1
[Arabidopsis thaliana] gi|21593673|gb|AAM65640.1|
En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 306
Score = 193 bits (491), Expect = 1e-48
Identities = 96/158 (60%), Positives = 113/158 (70%), Gaps = 12/158 (7%)
Frame = -2
Query: 619 LTPPSTPTDPGTPIDPGTP-------SIPSPPFFPAP-SPFTGTCNYWSTHPGIIWGILG 464
+TPPS DPGTPI G+P P PF PAP P TGTC+YW HP +IWG+LG
Sbjct: 148 VTPPSPIVDPGTPIIGGSPPTPIIDPGTPGTPFIPAPFPPITGTCDYWRNHPTLIWGLLG 207
Query: 463 WWGNLGSAFGV----TNAPGFGAGISLPQALSNTRSDGLGALYREGTASYLNSLVNSKFP 296
WWG +G AFG ++ PGF ++L QALSNTRSD +GALYREGTAS+LNS+VN KFP
Sbjct: 208 WWGTVGGAFGTVSIPSSIPGFDPHMNLLQALSNTRSDPIGALYREGTASWLNSMVNHKFP 267
Query: 295 FTTNQVREKFVASLHSNKAAAAQARLFKMANEGKMKPR 182
FTT QVR+ FVA L SNKAA QA FK+ANEG++KPR
Sbjct: 268 FTTPQVRDHFVAGLSSNKAATKQAHTFKLANEGRLKPR 305
Score = 33.1 bits (74), Expect = 3.0
Identities = 21/48 (43%), Positives = 25/48 (51%), Gaps = 7/48 (14%)
Frame = -2
Query: 610 PSTPTDPGTPIDPGTPSIPS---PPFFPA--PSPFTGTCNYWS--THP 488
PSTP+ P P TPS PS P P+ P+P T CN S +HP
Sbjct: 67 PSTPSHPSPPSHTPTPSTPSHTPTPHTPSHTPTPHTPPCNCGSPPSHP 114
>dbj|BAB90075.1| VsaA -like protein [Oryza sativa (japonica cultivar-group)]
Length = 647
Score = 157 bits (397), Expect = 1e-37
Identities = 79/142 (55%), Positives = 104/142 (72%), Gaps = 3/142 (2%)
Frame = -2
Query: 613 PPSTPTDPGTPIDPGTPSIPSP-PFFPAPSPFTGTCNYWSTHPGIIWGILGWWGNLGSAF 437
P STP+ P +P+ P PS +P PF P +PF C+YW +HPG+IWG+ G+W + F
Sbjct: 504 PISTPSSPYSPLVPTPPSSTTPMPFDPNTAPFP--CSYWLSHPGVIWGLFGFWCPMARLF 561
Query: 436 GVTNAPGFGAGISLPQALSNTRSDGLGALYREGTASYLNSLVNSKFPFTTNQVREKFVAS 257
G T A FG +++P+AL+NTR+DG+G LYREGTAS LNS+VNS+FPFTT QV++ F A+
Sbjct: 562 GPTAAAPFGHDLTVPEALANTRADGVGELYREGTASLLNSMVNSRFPFTTQQVKDAFSAA 621
Query: 256 LHS--NKAAAAQARLFKMANEG 197
L S + AAAAQA+LFK ANEG
Sbjct: 622 LSSGGDHAAAAQAQLFKKANEG 643
>dbj|BAB32984.1| P0489A05.4 [Oryza sativa (japonica cultivar-group)]
gi|20804531|dbj|BAB92225.1| B1015E06.26 [Oryza sativa
(japonica cultivar-group)]
Length = 601
Score = 110 bits (274), Expect = 2e-23
Identities = 67/152 (44%), Positives = 87/152 (57%), Gaps = 14/152 (9%)
Frame = -2
Query: 610 PSTPTDPGTPIDP----------GTPSIPSPPFFPAPSPFT-GTCNYWSTHPGIIWGILG 464
PSTP+ G+ P G P+ P+ P T GTC+YW +HP +W LG
Sbjct: 448 PSTPSGGGSSPTPSHGGGAYGGGGAPATPASHDGHGLIPTTPGTCDYWRSHPMEMWSALG 507
Query: 463 WW-GNLGSAFGVTNAPGFGAGISLPQALSNTRSDGLGALYREGTASYLNSLVNSKFPFTT 287
W ++G FG + + G G G+S+ AL+NTR DG G L REG A+ LNS+ S FP+T
Sbjct: 508 RWPSSVGHFFG-SGSGGAGTGMSIQDALANTRGDGAGELMREGAAALLNSMTRSGFPYTA 566
Query: 286 NQVREKFVASL--HSNKAAAAQARLFKMANEG 197
QVR+ F A+ S+ AAAAQA FK ANEG
Sbjct: 567 EQVRDAFAAAAGGGSDGAAAAQAAAFKKANEG 598
>ref|NP_115763.1| protein kinase, lysine deficient 4; putative protein kinase WNK4
[Homo sapiens] gi|15131540|emb|CAC48387.1|
serine/threonine protein kinase [Homo sapiens]
Length = 1231
Score = 44.7 bits (104), Expect = 0.001
Identities = 22/52 (42%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Frame = -2
Query: 622 SLTPPSTPTDPGTPIDPGTPSIP-------SPPFFPAPSPFTGTCNYWSTHP 488
S + P TP PG P PGTP P SPP P+PSPF+ + S++P
Sbjct: 791 SSSSPGTPLSPGNPFSPGTPISPGPIFPITSPPCHPSPSPFSPISSQVSSNP 842
>dbj|BAC04669.1| unnamed protein product [Homo sapiens]
Length = 620
Score = 44.7 bits (104), Expect = 0.001
Identities = 22/52 (42%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Frame = -2
Query: 622 SLTPPSTPTDPGTPIDPGTPSIP-------SPPFFPAPSPFTGTCNYWSTHP 488
S + P TP PG P PGTP P SPP P+PSPF+ + S++P
Sbjct: 258 SSSSPGTPLSPGNPFSPGTPISPGPIFPITSPPCHPSPSPFSPISSQVSSNP 309
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 602,137,556
Number of Sequences: 1393205
Number of extensions: 15410292
Number of successful extensions: 116708
Number of sequences better than 10.0: 697
Number of HSP's better than 10.0 without gapping: 56946
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 105543
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)