Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005206A_C02 KMC005206A_c02
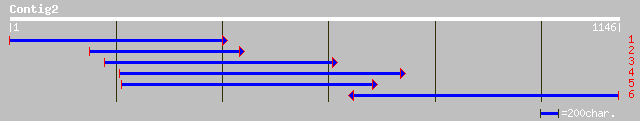
(1130 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC06126.1| putative ATP-dependent clp serine protease prote... 396 e-109
ref|NP_563893.1| ATP-dependent Clp protease proteolytic subunit ... 396 e-109
gb|AAB99906.1| ATP-dependent Clp protease proteolytic subunit [A... 390 e-107
pir||T07018 endopeptidase Clp (EC 3.4.21.92) chain P precursor, ... 378 e-103
gb|AAC04810.1| clp-like energy-dependent protease [Fritillaria a... 375 e-103
>emb|CAC06126.1| putative ATP-dependent clp serine protease proteolytic subunit [Pisum
sativum]
Length = 246
Score = 396 bits (1018), Expect = e-109
Identities = 199/246 (80%), Positives = 217/246 (87%)
Frame = -2
Query: 1129 SVPATATFSIASRRNTPSHLAFSHRNLCQIASNLSSPYGGHSSKFGLSSKSPGFPLKLDE 950
SVPA TF+IAS PS LAFSHRN C IAS+L SPY HSSK GLSSK+ GF LK DE
Sbjct: 1 SVPAFPTFTIASHTKIPSKLAFSHRNPCTIASSLPSPYV-HSSKIGLSSKAHGFRLKFDE 59
Query: 949 KNTYDASTSYGAIEAKKGNPPITPAVMTPGGPLDLSSVLFRNRIIFIGQPVNAQVAQRVI 770
KNT+D STSYG IEAKKGNPPI+PAVMTPGG +DLSSVLFRNRIIFIGQP+NAQ+AQRVI
Sbjct: 60 KNTHDVSTSYGVIEAKKGNPPISPAVMTPGGAIDLSSVLFRNRIIFIGQPINAQMAQRVI 119
Query: 769 SQLVTLATIDPNADILMYINCPGGSTYSVLAIYDCMSWIKPKVGTVCFGVAASQGALLLA 590
SQLVTLATIDP+ADILMYINC GGSTYSVLA+YDCMSWIKPKVGTVCFGVAASQ LLLA
Sbjct: 120 SQLVTLATIDPDADILMYINCAGGSTYSVLAVYDCMSWIKPKVGTVCFGVAASQATLLLA 179
Query: 589 GGEKGMRYSMPNSRIMIHQPQSGCGGHVEDVRRQVNEAVQSRHKIDKMYSAFTGQPIEKV 410
GGEKGMRYSMPN+RIM+ QP+ G GGHVEDV+R VNEAVQSRHK+D M+SAFTG PIE V
Sbjct: 180 GGEKGMRYSMPNARIMMGQPRCGFGGHVEDVKRLVNEAVQSRHKMDNMFSAFTGLPIETV 239
Query: 409 QQYTER 392
Q+YT+R
Sbjct: 240 QEYTDR 245
>ref|NP_563893.1| ATP-dependent Clp protease proteolytic subunit (ClpP6); protein id:
At1g11750.1, supported by cDNA: 33299., supported by
cDNA: gi_13194777, supported by cDNA: gi_2827887
[Arabidopsis thaliana] gi|25289851|pir||C86251
hypothetical protein [imported] - Arabidopsis thaliana
gi|4835782|gb|AAD30248.1|AC007296_9 Identical to
gb|AF016621 ATP-dependent Clp protease proteolytic
subunit from Arabidopsis thaliana and is a member of the
PF|00574 CLP_protease family. ESTs gb|F13836 and
gb|F13997 come from this gene
gi|13194778|gb|AAK15551.1|AF348580_1 putative
ATP-dependent Clp protease proteolytic subunit ClpP6
[Arabidopsis thaliana] gi|21592854|gb|AAM64804.1|
ATP-dependent Clp protease proteolytic subunit (ClpP6)
[Arabidopsis thaliana]
Length = 271
Score = 396 bits (1017), Expect = e-109
Identities = 203/271 (74%), Positives = 230/271 (83%), Gaps = 2/271 (0%)
Frame = -2
Query: 1129 SVPATATFSIASRRNTP-SHLAFSHRN-LCQIASNLSSPYGGHSSKFGLSSKSPGFPLKL 956
S P +FS +R P S L+ + RN + +I S L SPYG S K GLSS G P+K+
Sbjct: 7 SPPLGLSFSSRTRNPKPTSFLSHNQRNPIRRIVSALQSPYGD-SLKAGLSSNVSGSPIKI 65
Query: 955 DEKNTYDASTSYGAIEAKKGNPPITPAVMTPGGPLDLSSVLFRNRIIFIGQPVNAQVAQR 776
D K + +G IEAKKGNPP+ P+VMTPGGPLDLSSVLFRNRIIFIGQP+NAQVAQR
Sbjct: 66 DNK-----APRFGVIEAKKGNPPVMPSVMTPGGPLDLSSVLFRNRIIFIGQPINAQVAQR 120
Query: 775 VISQLVTLATIDPNADILMYINCPGGSTYSVLAIYDCMSWIKPKVGTVCFGVAASQGALL 596
VISQLVTLA+ID +DILMY+NCPGGSTYSVLAIYDCMSWIKPKVGTV FGVAASQGALL
Sbjct: 121 VISQLVTLASIDDKSDILMYLNCPGGSTYSVLAIYDCMSWIKPKVGTVAFGVAASQGALL 180
Query: 595 LAGGEKGMRYSMPNSRIMIHQPQSGCGGHVEDVRRQVNEAVQSRHKIDKMYSAFTGQPIE 416
LAGGEKGMRY+MPN+R+MIHQPQ+GCGGHVEDVRRQVNEA+++R KID+MY+AFTGQP+E
Sbjct: 181 LAGGEKGMRYAMPNTRVMIHQPQTGCGGHVEDVRRQVNEAIEARQKIDRMYAAFTGQPLE 240
Query: 415 KVQQYTERDRFLSVSEALEFGLIDGVLETEY 323
KVQQYTERDRFLS SEALEFGLIDG+LETEY
Sbjct: 241 KVQQYTERDRFLSASEALEFGLIDGLLETEY 271
>gb|AAB99906.1| ATP-dependent Clp protease proteolytic subunit [Arabidopsis thaliana]
Length = 271
Score = 390 bits (1001), Expect = e-107
Identities = 199/271 (73%), Positives = 226/271 (82%), Gaps = 2/271 (0%)
Frame = -2
Query: 1129 SVPATATFSIASRRNTP-SHLAFSHRN-LCQIASNLSSPYGGHSSKFGLSSKSPGFPLKL 956
S P +FS +R P S L+ + RN + +I S L SPYG S K GLSS G P+K+
Sbjct: 7 SPPLGLSFSSRTRNPKPTSFLSHNQRNPIRRIVSALQSPYGD-SLKAGLSSNVSGSPIKI 65
Query: 955 DEKNTYDASTSYGAIEAKKGNPPITPAVMTPGGPLDLSSVLFRNRIIFIGQPVNAQVAQR 776
D K + +G IEAKKGNPP+ P+VMTPGGPLDLSSVLFRNRIIFIGQP+NAQVAQR
Sbjct: 66 DNK-----APRFGVIEAKKGNPPVMPSVMTPGGPLDLSSVLFRNRIIFIGQPINAQVAQR 120
Query: 775 VISQLVTLATIDPNADILMYINCPGGSTYSVLAIYDCMSWIKPKVGTVCFGVAASQGALL 596
VISQLVTLA+ID +DILMY+NCPGGSTYSVL IYDCMSWIKPKVGTV FGVAASQGA
Sbjct: 121 VISQLVTLASIDDKSDILMYLNCPGGSTYSVLTIYDCMSWIKPKVGTVAFGVAASQGAFF 180
Query: 595 LAGGEKGMRYSMPNSRIMIHQPQSGCGGHVEDVRRQVNEAVQSRHKIDKMYSAFTGQPIE 416
LAGGEKGMRY+MPN+R+MIHQPQ+GCGGHVEDVRRQVNEA+++R KID+MY+AFTGQP+E
Sbjct: 181 LAGGEKGMRYAMPNTRVMIHQPQTGCGGHVEDVRRQVNEAIEARQKIDRMYAAFTGQPLE 240
Query: 415 KVQQYTERDRFLSVSEALEFGLIDGVLETEY 323
KVQQYTERDRFLS SEA EFGLIDG+LETEY
Sbjct: 241 KVQQYTERDRFLSASEAFEFGLIDGLLETEY 271
>pir||T07018 endopeptidase Clp (EC 3.4.21.92) chain P precursor, chloroplast
[similarity] - tomato gi|602758|gb|AAA81878.1| clp-like
energy-dependent protease
Length = 275
Score = 378 bits (970), Expect = e-103
Identities = 197/272 (72%), Positives = 224/272 (81%), Gaps = 3/272 (1%)
Frame = -2
Query: 1129 SVPATATFSIASRRNTPSHLAFSHRNLCQ-IASNLSSPYGGHSSKFGLSSKSPGFPLKLD 953
++ T+ ++SR T S R+L + + S L SPY SS G SSK G PLK +
Sbjct: 5 AIAGTSIVPVSSRHQTSFSSLLSSRSLRKNVVSVLRSPYSD-SSDIGFSSKKLGIPLKFN 63
Query: 952 E-KNTYDASTSYGAIEAKKG-NPPITPAVMTPGGPLDLSSVLFRNRIIFIGQPVNAQVAQ 779
E ++ ++SYG I AK+G NPPI PAVMTP G LDLS+VLFRNRIIFIGQPVN+ VAQ
Sbjct: 64 EYESGAHTNSSYGVIVAKEGANPPIMPAVMTPVGALDLSTVLFRNRIIFIGQPVNSAVAQ 123
Query: 778 RVISQLVTLATIDPNADILMYINCPGGSTYSVLAIYDCMSWIKPKVGTVCFGVAASQGAL 599
+VISQLVTLATID NADIL+Y+NCPGGSTYSVLAIYDCMSWIKPKVGTVCFG AASQGAL
Sbjct: 124 KVISQLVTLATIDENADILIYLNCPGGSTYSVLAIYDCMSWIKPKVGTVCFGAAASQGAL 183
Query: 598 LLAGGEKGMRYSMPNSRIMIHQPQSGCGGHVEDVRRQVNEAVQSRHKIDKMYSAFTGQPI 419
LLAGGEKGMRY+MPN+RIMIHQPQSGCGGHVEDVRRQVNEAVQSR K+DKMY+AFTGQ I
Sbjct: 184 LLAGGEKGMRYAMPNARIMIHQPQSGCGGHVEDVRRQVNEAVQSRQKVDKMYAAFTGQSI 243
Query: 418 EKVQQYTERDRFLSVSEALEFGLIDGVLETEY 323
E +Q YTERDRF+S +EA+EFGLIDGVLETEY
Sbjct: 244 EMIQTYTERDRFMSSAEAMEFGLIDGVLETEY 275
>gb|AAC04810.1| clp-like energy-dependent protease [Fritillaria agrestis]
Length = 267
Score = 375 bits (963), Expect = e-103
Identities = 184/225 (81%), Positives = 205/225 (90%), Gaps = 1/225 (0%)
Frame = -2
Query: 994 GLSSKSPGFPLKLDEKNTYDASTSYGA-IEAKKGNPPITPAVMTPGGPLDLSSVLFRNRI 818
G+ + PLKL + + +++ Y A IEAK+GNPP+ PAVMTPGGPLDLSSVLFRNRI
Sbjct: 43 GMFGGTNALPLKLTDPASCHSASRYDAVIEAKRGNPPMMPAVMTPGGPLDLSSVLFRNRI 102
Query: 817 IFIGQPVNAQVAQRVISQLVTLATIDPNADILMYINCPGGSTYSVLAIYDCMSWIKPKVG 638
IFIGQPVN+QVAQRVISQLVTLATID +DIL+Y+NCPGGSTYS+LAIYDCMSWIKPKVG
Sbjct: 103 IFIGQPVNSQVAQRVISQLVTLATIDEESDILVYLNCPGGSTYSILAIYDCMSWIKPKVG 162
Query: 637 TVCFGVAASQGALLLAGGEKGMRYSMPNSRIMIHQPQSGCGGHVEDVRRQVNEAVQSRHK 458
TVCFGVAASQGALLLAGGEKGMRY+MPN+RIMIHQPQSGCGGHVEDVRRQVNEAVQSRHK
Sbjct: 163 TVCFGVAASQGALLLAGGEKGMRYTMPNARIMIHQPQSGCGGHVEDVRRQVNEAVQSRHK 222
Query: 457 IDKMYSAFTGQPIEKVQQYTERDRFLSVSEALEFGLIDGVLETEY 323
ID+MY+AFTGQ +EKVQ+YTERDRF S SEALEFGL+DGVLETEY
Sbjct: 223 IDQMYAAFTGQTLEKVQEYTERDRFFSASEALEFGLVDGVLETEY 267
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 997,500,643
Number of Sequences: 1393205
Number of extensions: 22967498
Number of successful extensions: 62651
Number of sequences better than 10.0: 324
Number of HSP's better than 10.0 without gapping: 59589
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 62501
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 68909194122
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)