Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005183A_C01 KMC005183A_c01
(492 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
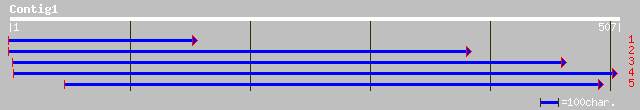
ref|NP_180571.1| putative unknown protein, leucine-rich repeat; ... 115 2e-25
ref|NP_196408.2| putative protein; protein id: At5g07910.1, supp... 60 1e-08
pir||T45616 hypothetical protein F13G24.110 - Arabidopsis thalia... 56 2e-07
sp|O93233|PLIB_AGKBL Phospholipase A2 inhibitor subunit B precur... 39 0.024
gb|EAA41265.1| GLP_190_17496_14935 [Giardia lamblia ATCC 50803] 35 0.45
>ref|NP_180571.1| putative unknown protein, leucine-rich repeat; protein id:
At2g30100.1 [Arabidopsis thaliana] gi|7445541|pir||T00588
hypothetical protein At2g30100 [imported] - Arabidopsis
thaliana gi|3150410|gb|AAC16962.1| putative unknown
protein, leucine-rich repeat [Arabidopsis thaliana]
Length = 902
Score = 115 bits (289), Expect = 2e-25
Identities = 54/78 (69%), Positives = 67/78 (85%)
Frame = -3
Query: 481 LKVLHLSNNGMKSLPSKLFKTCLQLSTLDLHNTEITIDILRQYEGWNNFDERRRSKHQKQ 302
++ L L+N G+K+LPS LFK CLQLSTL LHNTEIT++ LRQ+EG+++FDERRR+KHQKQ
Sbjct: 824 IQTLELNNTGLKTLPSALFKMCLQLSTLGLHNTEITVEFLRQFEGYDDFDERRRTKHQKQ 883
Query: 301 LDFRVGVSRDFDEGADKN 248
LDFRV S FDEGADK+
Sbjct: 884 LDFRVVGSGQFDEGADKS 901
>ref|NP_196408.2| putative protein; protein id: At5g07910.1, supported by cDNA:
gi_20260337 [Arabidopsis thaliana]
gi|20260338|gb|AAM13067.1| unknown protein [Arabidopsis
thaliana] gi|22136192|gb|AAM91174.1| unknown protein
[Arabidopsis thaliana]
Length = 262
Score = 60.1 bits (144), Expect = 1e-08
Identities = 32/78 (41%), Positives = 44/78 (56%), Gaps = 1/78 (1%)
Frame = -3
Query: 481 LKVLHLSNNGMKSLPSKLFKTCLQLSTLDLHNTEITIDILRQYEGWNNFDERRRSKHQKQ 302
LK L L NN + +P L C L L LHN I++D + EG+ +F+ERR+ K KQ
Sbjct: 185 LKSLSLDNNQVNQIPDGLLIHCKSLQNLSLHNNPISMDQFQLMEGYQDFEERRKKKFDKQ 244
Query: 301 LDFRVGV-SRDFDEGADK 251
+D V + S+ D G DK
Sbjct: 245 IDSNVMMGSKALDVGVDK 262
>pir||T45616 hypothetical protein F13G24.110 - Arabidopsis thaliana
gi|6562305|emb|CAB62603.1| putative protein [Arabidopsis
thaliana] gi|10176725|dbj|BAB09955.1|
gene_id:MXM12.15~unknown protein [Arabidopsis thaliana]
Length = 268
Score = 56.2 bits (134), Expect = 2e-07
Identities = 30/76 (39%), Positives = 43/76 (56%), Gaps = 1/76 (1%)
Frame = -3
Query: 481 LKVLHLSNNGMKSLPSKLFKTCLQLSTLDLHNTEITIDILRQYEGWNNFDERRRSKHQKQ 302
LK L L NN + +P L C L L LHN I++D + EG+ +F+ERR+ K KQ
Sbjct: 185 LKSLSLDNNQVNQIPDGLLIHCKSLQNLSLHNNPISMDQFQLMEGYQDFEERRKKKFDKQ 244
Query: 301 LDFRVGV-SRDFDEGA 257
+D V + S+ D G+
Sbjct: 245 IDSNVMMGSKALDVGS 260
>sp|O93233|PLIB_AGKBL Phospholipase A2 inhibitor subunit B precursor (PLI-B)
gi|3358089|dbj|BAA31994.1| phospholipase A2 inhibitor
[Agkistrodon blomhoffii siniticus]
Length = 331
Score = 39.3 bits (90), Expect = 0.024
Identities = 19/35 (54%), Positives = 24/35 (68%)
Frame = -3
Query: 481 LKVLHLSNNGMKSLPSKLFKTCLQLSTLDLHNTEI 377
L+ LHLSNN +K+LPS LF+ QL TLDL +
Sbjct: 81 LQELHLSNNRLKTLPSGLFRNLPQLHTLDLSTNHL 115
>gb|EAA41265.1| GLP_190_17496_14935 [Giardia lamblia ATCC 50803]
Length = 853
Score = 35.0 bits (79), Expect = 0.45
Identities = 17/38 (44%), Positives = 28/38 (72%)
Frame = -3
Query: 490 FSYLKVLHLSNNGMKSLPSKLFKTCLQLSTLDLHNTEI 377
F+ L+ ++LSNN ++S+PS +F +C +LS+ DL N I
Sbjct: 249 FTNLREINLSNNTLQSIPSFIF-SCPELSSADLSNNNI 285
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 384,102,729
Number of Sequences: 1393205
Number of extensions: 7248149
Number of successful extensions: 16696
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 15759
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 16694
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 14203329973
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)