Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005169A_C01 KMC005169A_c01
(945 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
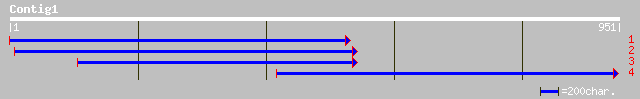
ref|NP_200384.1| fasciclin-like arabinogalactan-protein (FLA1); ... 304 e-104
gb|AAM65777.1| putative pollen surface protein [Arabidopsis thal... 255 7e-88
ref|NP_193009.1| fasciclin-like arabinogalactan-protein (FLA2); ... 255 7e-88
gb|AAK20858.1|AF333971_1 fasciclin-like arabinogalactan-protein ... 255 7e-88
dbj|BAC22390.1| putative fasciclin-like arabinogalactan-protein ... 217 1e-68
>ref|NP_200384.1| fasciclin-like arabinogalactan-protein (FLA1); protein id:
At5g55730.1, supported by cDNA: gi_13377775 [Arabidopsis
thaliana] gi|9758607|dbj|BAB09240.1|
gene_id:MDF20.17~pir||T06631~similar to unknown protein
[Arabidopsis thaliana]
gi|13377776|gb|AAK20857.1|AF333970_1 fasciclin-like
arabinogalactan-protein 1 [Arabidopsis thaliana]
gi|27311863|gb|AAO00897.1| putative protein [Arabidopsis
thaliana]
Length = 424
Score = 304 bits (778), Expect(2) = e-104
Identities = 149/231 (64%), Positives = 187/231 (80%)
Frame = +1
Query: 31 MAALAAALLLLTMASTTTNAHNITGILAKHPEFSTFNHYLTLTHLAAEINQRTTITVCAV 210
M++L +L + +T T+AHN+T +LA HP FS+F+H+LT THLA EIN+R TITVCAV
Sbjct: 5 MSSLIIIFNILLLLTTQTHAHNVTRLLANHPSFSSFSHFLTQTHLADEINRRRTITVCAV 64
Query: 211 NNAAMDDLLSKHPSITTVKNILSLHVLLDYFGAKKLHQITNGTALAATMYQATGTAPGSA 390
+NAAM L SK +++T+KNILSLHVLLDYFG KKLHQI +G+ALAAT++QATG APG++
Sbjct: 65 DNAAMSALTSKGYTLSTLKNILSLHVLLDYFGTKKLHQIRDGSALAATLFQATGAAPGTS 124
Query: 391 GFVNITDLRGGKVGFGAENNDGTLSASFVKSVEEIPYNISIIQISKVLPSAAAEAPAPAP 570
GFVNITDLRGGKVGFG + D LS+ FVKS+EE+PYNISIIQIS+VLPS A AP PAP
Sbjct: 125 GFVNITDLRGGKVGFGPDGGD--LSSFFVKSIEEVPYNISIIQISRVLPSETAAAPTPAP 182
Query: 571 AQQNLTAIMSKHGCKIFADTLSATPDAYSTFTDNLDGGLTVSCPVDDAVQG 723
A+ NLT IMS HGCK+FA+TL P A T+ ++L+GG+TV CP DDA++G
Sbjct: 183 AEMNLTGIMSAHGCKVFAETLLTNPGASKTYQESLEGGMTVFCPGDDAMKG 233
Score = 99.4 bits (246), Expect(2) = e-104
Identities = 50/81 (61%), Positives = 58/81 (70%)
Frame = +2
Query: 701 PSTTQFKASLPKFKNLTAAGKVSLLEFHAVPVYQSMATLKSRNGVQNTLATDGANKYDFT 880
P K LPK+KNLTA K + L+F AVP Y SMA LKS NG NTLATDGANK++ T
Sbjct: 226 PGDDAMKGFLPKYKNLTAPKKEAFLDFLAVPTYYSMAMLKSNNGPMNTLATDGANKFELT 285
Query: 881 VQNDGDKVTLKTSGVTARIID 943
VQNDG+KVTLKT T +I+D
Sbjct: 286 VQNDGEKVTLKTRINTVKIVD 306
>gb|AAM65777.1| putative pollen surface protein [Arabidopsis thaliana]
Length = 403
Score = 255 bits (652), Expect(2) = 7e-88
Identities = 131/229 (57%), Positives = 171/229 (74%), Gaps = 1/229 (0%)
Frame = +1
Query: 34 AALAAALLL-LTMASTTTNAHNITGILAKHPEFSTFNHYLTLTHLAAEINQRTTITVCAV 210
AA A L+ L + + +NAHNIT ILAK P+FSTFNHYL+ THLA EIN+R TITV AV
Sbjct: 7 AATALVLIFQLHLFLSLSNAHNITRILAKDPDFSTFNHYLSATHLADEINRRQTITVLAV 66
Query: 211 NNAAMDDLLSKHPSITTVKNILSLHVLLDYFGAKKLHQITNGTALAATMYQATGTAPGSA 390
+N+AM +LS S+ ++NILSLHVL+DYFG KKLHQIT+G+ A+M+Q+TG+A G++
Sbjct: 67 DNSAMSSILSNGYSLYQIRNILSLHVLVDYFGTKKLHQITDGSTSTASMFQSTGSATGTS 126
Query: 391 GFVNITDLRGGKVGFGAENNDGTLSASFVKSVEEIPYNISIIQISKVLPSAAAEAPAPAP 570
G++NITD++GGKV FG +++D L+A +VKSV E PYNIS++ IS+VL S AEAP +P
Sbjct: 127 GYINITDIKGGKVAFGVQDDDSKLTAHYVKSVFEKPYNISVLHISQVLTSPEAEAPTASP 186
Query: 571 AQQNLTAIMSKHGCKIFADTLSATPDAYSTFTDNLDGGLTVSCPVDDAV 717
+ LT I+ K GCK F+D L +T A TF D +DGGLTV CP D AV
Sbjct: 187 SDLILTTILEKQGCKAFSDILKST-GADKTFQDTVDGGLTVFCPSDSAV 234
Score = 92.0 bits (227), Expect(2) = 7e-88
Identities = 43/80 (53%), Positives = 60/80 (74%)
Frame = +2
Query: 701 PSTTQFKASLPKFKNLTAAGKVSLLEFHAVPVYQSMATLKSRNGVQNTLATDGANKYDFT 880
PS + +PKFK+L+ A K +L+ +H +PVYQS+ L+S NG NTLAT+G NK+DFT
Sbjct: 229 PSDSAVGKFMPKFKSLSPANKTALVLYHGMPVYQSLQMLRSGNGAVNTLATEGNNKFDFT 288
Query: 881 VQNDGDKVTLKTSGVTARII 940
VQNDG+ VTL+T VTA+++
Sbjct: 289 VQNDGEDVTLETDVVTAKVM 308
>ref|NP_193009.1| fasciclin-like arabinogalactan-protein (FLA2); protein id:
At4g12730.1, supported by cDNA: 4620., supported by
cDNA: gi_13377777, supported by cDNA: gi_16974608
[Arabidopsis thaliana] gi|7488019|pir||T06631 pollen
surface protein homolog T20K18.80 - Arabidopsis thaliana
gi|4586249|emb|CAB40990.1| putative pollen surface
protein [Arabidopsis thaliana]
gi|7267974|emb|CAB78315.1| putative pollen surface
protein [Arabidopsis thaliana]
gi|16974609|gb|AAL31207.1| AT4g12730/T20K18_80
[Arabidopsis thaliana] gi|22655474|gb|AAM98329.1|
At4g12730/T20K18_80 [Arabidopsis thaliana]
Length = 403
Score = 255 bits (652), Expect(2) = 7e-88
Identities = 131/229 (57%), Positives = 171/229 (74%), Gaps = 1/229 (0%)
Frame = +1
Query: 34 AALAAALLL-LTMASTTTNAHNITGILAKHPEFSTFNHYLTLTHLAAEINQRTTITVCAV 210
AA A L+ L + + +NAHNIT ILAK P+FSTFNHYL+ THLA EIN+R TITV AV
Sbjct: 7 AATALVLIFQLHLFLSLSNAHNITRILAKDPDFSTFNHYLSATHLADEINRRQTITVLAV 66
Query: 211 NNAAMDDLLSKHPSITTVKNILSLHVLLDYFGAKKLHQITNGTALAATMYQATGTAPGSA 390
+N+AM +LS S+ ++NILSLHVL+DYFG KKLHQIT+G+ A+M+Q+TG+A G++
Sbjct: 67 DNSAMSSILSNGYSLYQIRNILSLHVLVDYFGTKKLHQITDGSTSTASMFQSTGSATGTS 126
Query: 391 GFVNITDLRGGKVGFGAENNDGTLSASFVKSVEEIPYNISIIQISKVLPSAAAEAPAPAP 570
G++NITD++GGKV FG +++D L+A +VKSV E PYNIS++ IS+VL S AEAP +P
Sbjct: 127 GYINITDIKGGKVAFGVQDDDSKLTAHYVKSVFEKPYNISVLHISQVLTSPEAEAPTASP 186
Query: 571 AQQNLTAIMSKHGCKIFADTLSATPDAYSTFTDNLDGGLTVSCPVDDAV 717
+ LT I+ K GCK F+D L +T A TF D +DGGLTV CP D AV
Sbjct: 187 SDLILTTILEKQGCKAFSDILKST-GADKTFQDTVDGGLTVFCPSDSAV 234
Score = 92.0 bits (227), Expect(2) = 7e-88
Identities = 43/80 (53%), Positives = 60/80 (74%)
Frame = +2
Query: 701 PSTTQFKASLPKFKNLTAAGKVSLLEFHAVPVYQSMATLKSRNGVQNTLATDGANKYDFT 880
PS + +PKFK+L+ A K +L+ +H +PVYQS+ L+S NG NTLAT+G NK+DFT
Sbjct: 229 PSDSAVGKFMPKFKSLSPANKTALVLYHGMPVYQSLQMLRSGNGAVNTLATEGNNKFDFT 288
Query: 881 VQNDGDKVTLKTSGVTARII 940
VQNDG+ VTL+T VTA+++
Sbjct: 289 VQNDGEDVTLETDVVTAKVM 308
>gb|AAK20858.1|AF333971_1 fasciclin-like arabinogalactan-protein 2 [Arabidopsis thaliana]
Length = 403
Score = 255 bits (652), Expect(2) = 7e-88
Identities = 131/229 (57%), Positives = 171/229 (74%), Gaps = 1/229 (0%)
Frame = +1
Query: 34 AALAAALLL-LTMASTTTNAHNITGILAKHPEFSTFNHYLTLTHLAAEINQRTTITVCAV 210
AA A L+ L + + +NAHNIT ILAK P+FSTFNHYL+ THLA EIN+R TITV AV
Sbjct: 7 AATALVLIFQLHLFLSLSNAHNITRILAKDPDFSTFNHYLSATHLADEINRRQTITVLAV 66
Query: 211 NNAAMDDLLSKHPSITTVKNILSLHVLLDYFGAKKLHQITNGTALAATMYQATGTAPGSA 390
+N+AM +LS S+ ++NILSLHVL+DYFG KKLHQIT+G+ A+M+Q+TG+A G++
Sbjct: 67 DNSAMSSILSNGYSLYQIRNILSLHVLVDYFGTKKLHQITDGSTSTASMFQSTGSATGTS 126
Query: 391 GFVNITDLRGGKVGFGAENNDGTLSASFVKSVEEIPYNISIIQISKVLPSAAAEAPAPAP 570
G++NITD++GGKV FG +++D L+A +VKSV E PYNIS++ IS+VL S AEAP +P
Sbjct: 127 GYINITDIKGGKVAFGVQDDDSKLTAHYVKSVFEKPYNISVLHISQVLTSPEAEAPTASP 186
Query: 571 AQQNLTAIMSKHGCKIFADTLSATPDAYSTFTDNLDGGLTVSCPVDDAV 717
+ LT I+ K GCK F+D L +T A TF D +DGGLTV CP D AV
Sbjct: 187 SDLILTTILEKQGCKAFSDILKST-GADKTFQDTVDGGLTVFCPSDSAV 234
Score = 92.0 bits (227), Expect(2) = 7e-88
Identities = 43/80 (53%), Positives = 60/80 (74%)
Frame = +2
Query: 701 PSTTQFKASLPKFKNLTAAGKVSLLEFHAVPVYQSMATLKSRNGVQNTLATDGANKYDFT 880
PS + +PKFK+L+ A K +L+ +H +PVYQS+ L+S NG NTLAT+G NK+DFT
Sbjct: 229 PSDSAVGKFMPKFKSLSPANKTALVLYHGMPVYQSLQMLRSGNGAVNTLATEGNNKFDFT 288
Query: 881 VQNDGDKVTLKTSGVTARII 940
VQNDG+ VTL+T VTA+++
Sbjct: 289 VQNDGEDVTLETDVVTAKVM 308
>dbj|BAC22390.1| putative fasciclin-like arabinogalactan-protein [Oryza sativa
(japonica cultivar-group)]
Length = 459
Score = 217 bits (552), Expect(2) = 1e-68
Identities = 117/240 (48%), Positives = 158/240 (65%), Gaps = 1/240 (0%)
Frame = +1
Query: 1 STMQLLRPATMAALAAALLLLTMASTTTNAHNITGILAKHPEFSTFNHYLTLTHLAAEIN 180
S M+LL A+ A++ LT A+T +NIT IL HPE+S FN LT T LA +IN
Sbjct: 43 SNMELL--LRRLAVVVAVVALT-AATAAEGYNITKILGDHPEYSQFNKLLTETRLAGDIN 99
Query: 181 QRTTITVCAVNNAAMDDLLSKHPSITTVKNILSLHVLLDYFGAKKLHQITNGTALAATMY 360
+R TITV V N M L H ++ T+++IL +H+L+DY+GAKKLHQ+ G +++M+
Sbjct: 100 RRRTITVLVVANGDMGALSGGHYTLPTLRHILEMHILVDYYGAKKLHQLARGDTASSSMF 159
Query: 361 QATGTAPGSAGFVNITDLRGGKVGFGAEN-NDGTLSASFVKSVEEIPYNISIIQISKVLP 537
Q +G+APG+ G+VNIT RGG+V F AE+ D +SFVKSV+EIPY+++++QISK L
Sbjct: 160 QESGSAPGTTGYVNITQHRGGRVSFTAEDAADSATPSSFVKSVKEIPYDLAVLQISKPLS 219
Query: 538 SAAAEAPAPAPAQQNLTAIMSKHGCKIFADTLSATPDAYSTFTDNLDGGLTVSCPVDDAV 717
S AEAP PA NLT ++SK CK FA L++ D YS D GLT+ CPVD AV
Sbjct: 220 SPEAEAPVAPPAPVNLTELLSKKYCKNFAGLLASNADVYSNINATKDNGLTLFCPVDAAV 279
Score = 66.2 bits (160), Expect(2) = 1e-68
Identities = 40/87 (45%), Positives = 50/87 (56%), Gaps = 6/87 (6%)
Frame = +2
Query: 701 PSTTQFKASLPKFKNLTAAGKVSLLEFHAVPVYQSMATLKSRNGVQNTLATDGANK--YD 874
P A LPK+KNLTA GK ++L +HAVP Y S+ LKS +G +TLAT K Y
Sbjct: 274 PVDAAVDAFLPKYKNLTAKGKAAILLYHAVPDYYSLQLLKSNSGKVSTLATASVAKKDYS 333
Query: 875 FTVQNDGDKVTLKT----SGVTARIID 943
+ V ND D V L T + VTA + D
Sbjct: 334 YDVSNDRDSVLLDTKVNSASVTATVKD 360
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.317 0.130 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,007,481,105
Number of Sequences: 1393205
Number of extensions: 30477103
Number of successful extensions: 664202
Number of sequences better than 10.0: 21174
Number of HSP's better than 10.0 without gapping: 171478
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 395382
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 52969081112
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)