Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005167A_C01 KMC005167A_c01
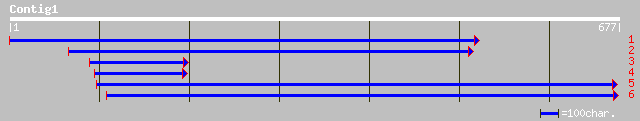
(673 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC69138.1| MAP kinase kinase [Medicago sativa subsp. x varia] 201 7e-51
gb|AAF67262.1|AF165186_1 MAP kinase kinase [Nicotiana tabacum] 166 2e-40
emb|CAA04261.2| MAP kinase kinase [Lycopersicon esculentum] 157 2e-37
pir||T06583 protein kinase MEK1 - tomato 157 2e-37
ref|NP_194710.1| MAP kinase kinase 2; protein id: At4g29810.1, s... 152 4e-36
>emb|CAC69138.1| MAP kinase kinase [Medicago sativa subsp. x varia]
Length = 356
Score = 201 bits (511), Expect = 7e-51
Identities = 97/107 (90%), Positives = 100/107 (92%)
Frame = -2
Query: 672 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 493
SDIWSLGLILLECAMGRFPY PPDQSE WESIFELIET+VDKPPPSAPSEQFS+EFCSFI
Sbjct: 250 SDIWSLGLILLECAMGRFPYTPPDQSERWESIFELIETIVDKPPPSAPSEQFSSEFCSFI 309
Query: 492 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSPLATL 352
SACLQKDP RLSAQELM LPF SMYDDL VDLSAYFS+AGSPLATL
Sbjct: 310 SACLQKDPGSRLSAQELMELPFISMYDDLHVDLSAYFSDAGSPLATL 356
>gb|AAF67262.1|AF165186_1 MAP kinase kinase [Nicotiana tabacum]
Length = 357
Score = 166 bits (421), Expect = 2e-40
Identities = 75/107 (70%), Positives = 91/107 (84%)
Frame = -2
Query: 672 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 493
SDIWSLGL+LLECA G FPY+PP E W +++EL+ET+VD+P PSAP +QFS +FCSFI
Sbjct: 248 SDIWSLGLVLLECATGVFPYSPPQADEGWVNVYELMETIVDQPAPSAPPDQFSPQFCSFI 307
Query: 492 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSPLATL 352
SAC+QKD KDRLSA ELMR PF +MYDDLD+DL +YF++AG PLATL
Sbjct: 308 SACVQKDQKDRLSANELMRHPFITMYDDLDIDLGSYFTSAGPPLATL 354
>emb|CAA04261.2| MAP kinase kinase [Lycopersicon esculentum]
Length = 357
Score = 157 bits (396), Expect = 2e-37
Identities = 70/107 (65%), Positives = 87/107 (80%)
Frame = -2
Query: 672 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 493
SDIWSLGL+LLECA G FPY PP+ E W +++EL+ET+VD+P P AP +QFS +FCSFI
Sbjct: 248 SDIWSLGLVLLECATGHFPYKPPEGDEGWVNVYELMETIVDQPEPCAPPDQFSPQFCSFI 307
Query: 492 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSPLATL 352
SAC+QK KDRLSA +LM PF +MYDD D+DL +YF++AG PLATL
Sbjct: 308 SACVQKHQKDRLSANDLMSHPFITMYDDQDIDLGSYFTSAGPPLATL 354
>pir||T06583 protein kinase MEK1 - tomato
Length = 357
Score = 157 bits (396), Expect = 2e-37
Identities = 70/107 (65%), Positives = 87/107 (80%)
Frame = -2
Query: 672 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 493
SDIWSLGL+LLECA G FPY PP+ E W +++EL+ET+VD+P P AP +QFS +FCSFI
Sbjct: 248 SDIWSLGLVLLECATGHFPYKPPEGDEGWVNVYELMETIVDQPEPCAPPDQFSPQFCSFI 307
Query: 492 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSPLATL 352
SAC+QK KDRLSA +LM PF +MYDD D+DL +YF++AG PLATL
Sbjct: 308 SACVQKHQKDRLSANDLMSHPFITMYDDQDIDLGSYFTSAGPPLATL 354
>ref|NP_194710.1| MAP kinase kinase 2; protein id: At4g29810.1, supported by cDNA:
gi_14326470, supported by cDNA: gi_18700199, supported
by cDNA: gi_3219266, supported by cDNA: gi_3859485
[Arabidopsis thaliana] gi|7434321|pir||T08542
mitogen-activated protein kinase kinase (EC 2.7.1.-) 2
[similarity] - Arabidopsis thaliana
gi|3219267|dbj|BAA28828.1| MAP kinase kinase 2
[Arabidopsis thaliana] gi|4914405|emb|CAB43656.1| MAP
kinase kinase 2 [Arabidopsis thaliana]
gi|7269880|emb|CAB79739.1| MAP kinase kinase 2
[Arabidopsis thaliana]
gi|14326471|gb|AAK60281.1|AF385688_1 AT4g29810/F27B13_50
[Arabidopsis thaliana] gi|18700200|gb|AAL77710.1|
AT4g29810/F27B13_50 [Arabidopsis thaliana]
Length = 363
Score = 152 bits (384), Expect = 4e-36
Identities = 69/107 (64%), Positives = 86/107 (79%)
Frame = -2
Query: 672 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 493
SDIWSLGL++LECA G+FPYAPP+Q ETW S+FEL+E +VD+PPP+ PS FS E SFI
Sbjct: 248 SDIWSLGLVVLECATGKFPYAPPNQEETWTSVFELMEAIVDQPPPALPSGNFSPELSSFI 307
Query: 492 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSPLATL 352
S CLQKDP R SA+ELM PF + YD ++L++YF++AGSPLATL
Sbjct: 308 STCLQKDPNSRSSAKELMEHPFLNKYDYSGINLASYFTDAGSPLATL 354
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 554,101,413
Number of Sequences: 1393205
Number of extensions: 11500554
Number of successful extensions: 31724
Number of sequences better than 10.0: 1807
Number of HSP's better than 10.0 without gapping: 29908
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30905
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29421376608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)