Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005159A_C01 KMC005159A_c01
(925 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
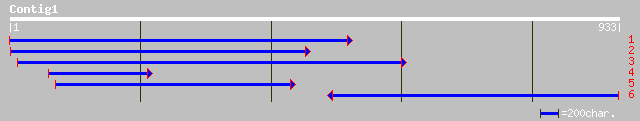
pir||T05720 ribosomal protein L22, chloroplast - soybean (fragme... 159 1e-52
sp|P49163|RK22_MEDSA 50S RIBOSOMAL PROTEIN L22, CHLOROPLAST PREC... 160 1e-51
sp|P23408|RK22_PEA 50S RIBOSOMAL PROTEIN L22, CHLOROPLAST PRECUR... 160 9e-50
ref|NP_783270.1| ribosomal protein L22 [Atropa belladonna] gi|20... 111 2e-30
ref|NP_054538.1| ribosomal protein L22 [Nicotiana tabacum] gi|13... 108 5e-30
>pir||T05720 ribosomal protein L22, chloroplast - soybean (fragment)
gi|2738560|gb|AAB94504.1| ribosomal protein L22 [Glycine
max]
Length = 164
Score = 159 bits (402), Expect(2) = 1e-52
Identities = 86/121 (71%), Positives = 97/121 (80%)
Frame = -3
Query: 773 CSSSPFTDKTLISLQPTTVSNPNNPFVCHARRARAPFGGQETNQQYVEAYAVGRHIRTSA 594
CSSSPF DKTLIS+ P T NP C ++ FG QE+N+ YVEAYAVGR+IR SA
Sbjct: 1 CSSSPFNDKTLISITPNTSHLNPNPLACRFHASQ--FGEQESNK-YVEAYAVGRNIRMSA 57
Query: 593 DKARRVIDQIRGRPYEETLTILELMPYRACEPILNIVFSAGANASHHLGLSKSSLVISKA 414
+KARRVIDQIRGR Y+ETL +LELMPYRACE IL IVFSAGANAS++LGLSK SLVISKA
Sbjct: 58 NKARRVIDQIRGRKYDETLMVLELMPYRACEAILKIVFSAGANASNNLGLSKGSLVISKA 117
Query: 413 E 411
E
Sbjct: 118 E 118
Score = 70.5 bits (171), Expect(2) = 1e-52
Identities = 33/52 (63%), Positives = 42/52 (80%)
Frame = -1
Query: 427 LLVKQSVNEGKTMKRMRPRAQGRAYQIKKRSCHIAIPVTGVPSDTAVEAKAA 272
++ K VNEGKTMKR++P A+GRA QIKKR+CHI I V G+PS++ VEAK A
Sbjct: 113 VISKAEVNEGKTMKRVKPVARGRAIQIKKRTCHITITVKGLPSESVVEAKPA 164
>sp|P49163|RK22_MEDSA 50S RIBOSOMAL PROTEIN L22, CHLOROPLAST PRECURSOR (CL22)
gi|7441002|pir||T09389 ribosomal protein L22,
chloroplast - alfalfa gi|552436|gb|AAB46612.1| ribosomal
protein CL22 [Medicago sativa]
Length = 200
Score = 160 bits (404), Expect(2) = 1e-51
Identities = 99/162 (61%), Positives = 111/162 (68%), Gaps = 8/162 (4%)
Frame = -3
Query: 872 MAVSF-AVNRP-------LLPSPLAPSQPKLKFPSRSFAIRCSSSPFTDKTLISLQPTTV 717
MA+S A+N P LL S LKFP I SSS F +L ++ P
Sbjct: 1 MALSLTAINLPPPPLRDNLLSSQFQFRPNLLKFPK----IPSSSSSFNGISLKTVTPN-- 54
Query: 716 SNPNNPFVCHARRARAPFGGQETNQQYVEAYAVGRHIRTSADKARRVIDQIRGRPYEETL 537
+N NNPF CH ++ FG QETN+ Y EA AVG+HIR SADKARRVID IRGRPYEETL
Sbjct: 55 NNNNNPFACHVSTSQ--FGVQETNKSYAEAVAVGKHIRMSADKARRVIDTIRGRPYEETL 112
Query: 536 TILELMPYRACEPILNIVFSAGANASHHLGLSKSSLVISKAE 411
ILELMPYRACE IL IVFSAGANAS++LGLSKSSLVISKAE
Sbjct: 113 MILELMPYRACETILKIVFSAGANASNNLGLSKSSLVISKAE 154
Score = 66.2 bits (160), Expect(2) = 1e-51
Identities = 30/52 (57%), Positives = 41/52 (78%)
Frame = -1
Query: 427 LLVKQSVNEGKTMKRMRPRAQGRAYQIKKRSCHIAIPVTGVPSDTAVEAKAA 272
++ K VNEG+TMKR RPRAQGRA +I KR+CHI I V G+P+++ VEA ++
Sbjct: 149 VISKAEVNEGRTMKRTRPRAQGRANRILKRTCHITITVKGLPAESVVEASSS 200
>sp|P23408|RK22_PEA 50S RIBOSOMAL PROTEIN L22, CHLOROPLAST PRECURSOR (CL22)
gi|100064|pir||S17314 ribosomal protein L22 precursor,
chloroplast - garden pea gi|169064|gb|AAA33656.1|
ribosomal protein CL22 gi|169066|gb|AAA33657.1|
ribosomal protein CL22
Length = 204
Score = 160 bits (405), Expect(2) = 9e-50
Identities = 100/164 (60%), Positives = 111/164 (66%), Gaps = 10/164 (6%)
Frame = -3
Query: 872 MAVSFAVNRPLLPSPLAPSQP--------KLKFPSRSFA-IRCSSSP-FTDKTLISLQPT 723
MA+S A+N P + L QP LKFP S A IRCSSS F D ISL+
Sbjct: 1 MALSLAINVPPVRDNLLRPQPFQSQFRPNLLKFPIPSSARIRCSSSSSFND---ISLKTV 57
Query: 722 TVSNPNNPFVCHARRARAPFGGQETNQQYVEAYAVGRHIRTSADKARRVIDQIRGRPYEE 543
T N N+ FVC + QETNQ Y E YAVGRHIR SADKARRV+DQIRGRPYE
Sbjct: 58 TPDNKNS-FVCRFNATQLEV--QETNQPYAETYAVGRHIRMSADKARRVVDQIRGRPYEA 114
Query: 542 TLTILELMPYRACEPILNIVFSAGANASHHLGLSKSSLVISKAE 411
+L +LELMPYRACE I+ IVFSAGANASH+LGLSKSSL ISKAE
Sbjct: 115 SLMVLELMPYRACEAIIKIVFSAGANASHNLGLSKSSLFISKAE 158
Score = 59.7 bits (143), Expect(2) = 9e-50
Identities = 28/49 (57%), Positives = 36/49 (73%)
Frame = -1
Query: 418 KQSVNEGKTMKRMRPRAQGRAYQIKKRSCHIAIPVTGVPSDTAVEAKAA 272
K VNEGKT+KR+R RAQGRA QI KR+CHI I V G+P ++ E ++
Sbjct: 156 KAEVNEGKTLKRVRARAQGRANQILKRTCHITITVRGLPDESDKEENSS 204
>ref|NP_783270.1| ribosomal protein L22 [Atropa belladonna]
gi|20068370|emb|CAC88083.1| ribosomal protein L22
[Atropa belladonna]
Length = 155
Score = 111 bits (277), Expect(2) = 2e-30
Identities = 55/74 (74%), Positives = 64/74 (86%)
Frame = -3
Query: 632 EAYAVGRHIRTSADKARRVIDQIRGRPYEETLTILELMPYRACEPILNIVFSAGANASHH 453
E YA+G HI SADKARRVIDQIRGR YEETL ILELMPYRAC PIL +V+SA ANAS++
Sbjct: 8 EVYALGEHISMSADKARRVIDQIRGRSYEETLMILELMPYRACYPILKLVYSAAANASYN 67
Query: 452 LGLSKSSLVISKAE 411
+G S+++LVISKAE
Sbjct: 68 MGSSEANLVISKAE 81
Score = 43.9 bits (102), Expect(2) = 2e-30
Identities = 17/36 (47%), Positives = 28/36 (77%)
Frame = -1
Query: 427 LLVKQSVNEGKTMKRMRPRAQGRAYQIKKRSCHIAI 320
++ K VN G T+K+++PRA+GR++ IK+ +CHI I
Sbjct: 76 VISKAEVNGGTTVKKLKPRARGRSFPIKRSTCHITI 111
>ref|NP_054538.1| ribosomal protein L22 [Nicotiana tabacum]
gi|132794|sp|P06389|RK22_TOBAC Chloroplast 50S ribosomal
protein L22 gi|71288|pir||R5NT22 ribosomal protein L22 -
common tobacco chloroplast gi|11866|emb|CAA77382.1|
ribosomal protein L22 [Nicotiana tabacum]
gi|225236|prf||1211235BU ribosomal protein L22
Length = 155
Score = 108 bits (271), Expect(2) = 5e-30
Identities = 53/74 (71%), Positives = 64/74 (85%)
Frame = -3
Query: 632 EAYAVGRHIRTSADKARRVIDQIRGRPYEETLTILELMPYRACEPILNIVFSAGANASHH 453
E YA+G HI SADKARRVI+QIRGR YEETL ILELMPYRAC PIL +++SA ANAS++
Sbjct: 8 EVYALGEHISMSADKARRVINQIRGRSYEETLMILELMPYRACYPILKLIYSAAANASYN 67
Query: 452 LGLSKSSLVISKAE 411
+G S+++LVISKAE
Sbjct: 68 MGSSEANLVISKAE 81
Score = 45.1 bits (105), Expect(2) = 5e-30
Identities = 18/44 (40%), Positives = 31/44 (69%)
Frame = -1
Query: 427 LLVKQSVNEGKTMKRMRPRAQGRAYQIKKRSCHIAIPVTGVPSD 296
++ K VN G T+K+++PRA+GR++ IK+ +CHI I + + D
Sbjct: 76 VISKAEVNGGTTVKKLKPRARGRSFPIKRSTCHITIVMKDISLD 119
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 841,064,785
Number of Sequences: 1393205
Number of extensions: 20755311
Number of successful extensions: 70109
Number of sequences better than 10.0: 254
Number of HSP's better than 10.0 without gapping: 60692
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 68964
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51027805888
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)