Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005146A_C01 KMC005146A_c01
(663 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
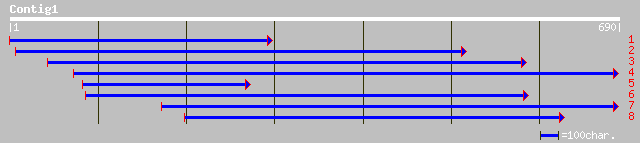
dbj|BAA96146.1| unnamed protein product [Oryza sativa (japonica ... 166 3e-40
ref|NP_173153.1| SOUL-like protein; protein id: At1g17100.1, sup... 159 2e-38
ref|NP_565181.1| expressed protein; protein id: At1g78450.1, sup... 119 4e-26
gb|AAM65865.1| SOUL-like protein [Arabidopsis thaliana] 119 5e-26
dbj|BAA92902.1| unnamed protein product [Oryza sativa (japonica ... 87 3e-16
>dbj|BAA96146.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|8096616|dbj|BAA96188.1| unnamed protein product
[Oryza sativa (japonica cultivar-group)]
Length = 218
Score = 166 bits (420), Expect = 3e-40
Identities = 79/125 (63%), Positives = 98/125 (78%), Gaps = 1/125 (0%)
Frame = -1
Query: 663 EMTAPVLSEVSPSDGPFCESSFVVSFFVPKVNLANPPPAKGLHVQRWKPVN-VAVRQFGG 487
EMTAPVL++V+PSDGPFC SSFVVSF+VP N +PPPA+GLHVQRW AVR+FGG
Sbjct: 90 EMTAPVLTQVAPSDGPFCVSSFVVSFYVPAKNQPDPPPAEGLHVQRWAGARYAAVRRFGG 149
Query: 486 FVSDASVGEEAAALKASIAGTKWAAALEKSHRAGHASVYSVAQYHAPFEYDNRVNEIWFL 307
FV+D+ VGE+AA L AS+ GT+WAAA+ RA S Y+VAQY++PFE+ RVNEIW L
Sbjct: 150 FVADSDVGEQAALLDASLQGTRWAAAVSDGRRADPTSSYTVAQYNSPFEFSGRVNEIWML 209
Query: 306 FDVEN 292
FD ++
Sbjct: 210 FDAKD 214
>ref|NP_173153.1| SOUL-like protein; protein id: At1g17100.1, supported by cDNA:
29992. [Arabidopsis thaliana] gi|25372687|pir||G86306
Similar to SOUL Protein [imported] - Arabidopsis
thaliana gi|5734756|gb|AAD50021.1|AC007651_16 Similar to
SOUL Protein [Arabidopsis thaliana]
gi|21592576|gb|AAM64525.1| SOUL-like protein
[Arabidopsis thaliana]
Length = 232
Score = 159 bits (403), Expect = 2e-38
Identities = 76/126 (60%), Positives = 96/126 (75%), Gaps = 3/126 (2%)
Frame = -1
Query: 663 EMTAPVLSEVSPSDGPFCESSFVVSFFVPKVNLANPPPAKGLHVQRWKPVNVAVRQFGGF 484
EMTAPV+S+VSPSDGPFCESSF VSF+VPK N +P P++ LH+Q+W VAVRQF GF
Sbjct: 106 EMTAPVISQVSPSDGPFCESSFTVSFYVPKKNQPDPAPSENLHIQKWNSRYVAVRQFSGF 165
Query: 483 VSDASVGEEAAALKASIAGTKWAAALEKSHR---AGHASVYSVAQYHAPFEYDNRVNEIW 313
VSD S+GE+AAAL +S+ GT WA A+ KS G S Y+VAQY++PFE+ RVNEIW
Sbjct: 166 VSDDSIGEQAAALDSSLKGTAWANAIAKSKEDGGVGSDSAYTVAQYNSPFEFSGRVNEIW 225
Query: 312 FLFDVE 295
F+++
Sbjct: 226 LPFELD 231
>ref|NP_565181.1| expressed protein; protein id: At1g78450.1, supported by cDNA:
5677. [Arabidopsis thaliana] gi|25372685|pir||H96812
hypothetical protein T30F21.21 [imported] - Arabidopsis
thaliana gi|4836887|gb|AAD30590.1|AC007260_21
Hypothetical protein [Arabidopsis thaliana]
gi|8052530|gb|AAF71794.1|AC013430_3 F3F9.4 [Arabidopsis
thaliana]
Length = 225
Score = 119 bits (298), Expect = 4e-26
Identities = 60/128 (46%), Positives = 84/128 (64%), Gaps = 3/128 (2%)
Frame = -1
Query: 663 EMTAPVLSEVSPSDGPFCESSFVVSFFVPKVNLANPPPAKGLHVQRWKPVNVAVRQFGGF 484
E+ P +++VS + S+F+VSFFVPK +PPP LHVQRW VAV+Q G+
Sbjct: 95 EIALPYITQVSQN-----LSTFIVSFFVPKAFQPDPPPGNNLHVQRWDSRYVAVKQISGY 149
Query: 483 VSDASVGEEAAALKASIAGTKWAAALEKSHR---AGHASVYSVAQYHAPFEYDNRVNEIW 313
V+D +G++ A LKAS+ GT WA A+EKS G A Y+VAQ+ PF++ RVNEIW
Sbjct: 150 VADHKIGKQVAELKASLQGTVWAKAIEKSRETGGVGSAWAYTVAQFSWPFQWSQRVNEIW 209
Query: 312 FLFDVEND 289
F F++E++
Sbjct: 210 FPFEMEDE 217
>gb|AAM65865.1| SOUL-like protein [Arabidopsis thaliana]
Length = 225
Score = 119 bits (297), Expect = 5e-26
Identities = 60/128 (46%), Positives = 84/128 (64%), Gaps = 3/128 (2%)
Frame = -1
Query: 663 EMTAPVLSEVSPSDGPFCESSFVVSFFVPKVNLANPPPAKGLHVQRWKPVNVAVRQFGGF 484
E+ P +++VS + S+F+VSFFVPK +PPP LHVQRW VAV+Q G+
Sbjct: 95 EIALPYITQVSQN-----LSTFIVSFFVPKAFQPDPPPGNNLHVQRWDSRYVAVKQISGY 149
Query: 483 VSDASVGEEAAALKASIAGTKWAAALEKSHRAG---HASVYSVAQYHAPFEYDNRVNEIW 313
V+D +G++ A LKAS+ GT WA A+EKS G A Y+VAQ+ PF++ RVNEIW
Sbjct: 150 VADHRIGKQVAELKASLQGTVWAKAIEKSRETGGVRSAWAYTVAQFSWPFQWSQRVNEIW 209
Query: 312 FLFDVEND 289
F F++E++
Sbjct: 210 FPFEMEDE 217
>dbj|BAA92902.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|8468007|dbj|BAA96608.1| unnamed protein product
[Oryza sativa (japonica cultivar-group)]
Length = 214
Score = 86.7 bits (213), Expect = 3e-16
Identities = 47/126 (37%), Positives = 67/126 (52%), Gaps = 3/126 (2%)
Frame = -1
Query: 660 MTAPVLSEVSPSDGPFCESSFVVSFFVPKVNLANPP---PAKGLHVQRWKPVNVAVRQFG 490
MT P+L+ + P GP S++ V ++P A+PP P LH RW +AVR F
Sbjct: 87 MTTPILTSIVPGAGPLHSSAYFVRLYLPAKFQASPPVPLPELNLHPDRWPSHCIAVRSFS 146
Query: 489 GFVSDASVGEEAAALKASIAGTKWAAALEKSHRAGHASVYSVAQYHAPFEYDNRVNEIWF 310
G+ D +V EEA L S++ + WA S S YS+AQY+ PF R+NE+WF
Sbjct: 147 GYARDNNVVEEAEKLALSLSRSPWA----NSTNYPSKSAYSIAQYNNPFRIIGRLNEVWF 202
Query: 309 LFDVEN 292
D ++
Sbjct: 203 DVDCKS 208
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 596,507,415
Number of Sequences: 1393205
Number of extensions: 13286989
Number of successful extensions: 31937
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 30126
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31752
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28572683052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)