Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005137A_C01 KMC005137A_c01
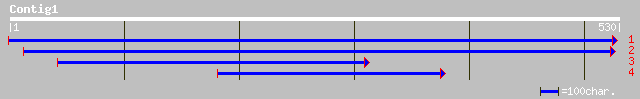
(528 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_188317.1| glycosyl hydrolase family 19 (chitinase); prote... 252 2e-66
ref|NP_172076.1| glycosyl hydrolase family 19 (chitinase); prote... 237 6e-62
dbj|BAC55635.1| OJ1081_B12.16 [Oryza sativa (japonica cultivar-g... 204 4e-52
gb|AAF69789.1|AF135149_1 class I chitinase [Arabis microphylla] 92 3e-18
gb|AAF69774.1|AF135134_1 class I chitinase [Arabis blepharophylla] 91 1e-17
>ref|NP_188317.1| glycosyl hydrolase family 19 (chitinase); protein id: At3g16920.1,
supported by cDNA: gi_15724261, supported by cDNA:
gi_19699111 [Arabidopsis thaliana]
gi|7670022|dbj|BAA94976.1| basic chitinase [Arabidopsis
thaliana] gi|15724262|gb|AAL06524.1|AF412071_1
AT3g16920/K14A17_4 [Arabidopsis thaliana]
gi|19699112|gb|AAL90922.1| AT3g16920/K14A17_4
[Arabidopsis thaliana]
Length = 333
Score = 252 bits (643), Expect = 2e-66
Identities = 115/167 (68%), Positives = 130/167 (76%), Gaps = 6/167 (3%)
Frame = +2
Query: 44 VLTTLVIFLAVSSTVEGDDNT------KTLVKYKHGKKYCDKGWECKGWSIYCCNLTISD 205
+LT ++ + V +D+ K LVK GKK CDKGWECKGWS YCCN TISD
Sbjct: 11 LLTVALVVFQTGTLVNAEDSEPSSSTRKPLVKIVKGKKLCDKGWECKGWSEYCCNHTISD 70
Query: 206 YFQVYQFENLFSKRNTPVANAVGFWDYHSFITAAALFEPQGFGTTGNKTLQMMEIAAFLG 385
+F+ YQFENLFSKRN+PVA+AVGFWDY SFITAAA ++P GFGT G K M E+AAFLG
Sbjct: 71 FFETYQFENLFSKRNSPVAHAVGFWDYRSFITAAAEYQPLGFGTAGEKLQGMKEVAAFLG 130
Query: 386 HVGAKTSCGYGVATGGPLAWGLCYNHEMSPSQKYCDEYYKLTYPCTP 526
HVG+KTSCGYGVATGGPLAWGLCYN EMSP Q YCD+YYKLTYPCTP
Sbjct: 131 HVGSKTSCGYGVATGGPLAWGLCYNKEMSPDQLYCDDYYKLTYPCTP 177
>ref|NP_172076.1| glycosyl hydrolase family 19 (chitinase); protein id: At1g05850.1,
supported by cDNA: gi_12083323, supported by cDNA:
gi_14334487, supported by cDNA: gi_17226328, supported
by cDNA: gi_21280934 [Arabidopsis thaliana]
gi|25406872|pir||C86193 hypothetical protein [imported]
- Arabidopsis thaliana
gi|6850314|gb|AAF29391.1|AC009999_11 Contains similarity
to a basic endochitinase from Arabidopis thaliana
gb|AB023448, and contains a Chitinases class I PF|00182
domain. ESTs gb|AI995747, gb|AA728545, gb|Z26222,
gb|Z25683, gb|T88386, gb|T14122, gb|T04241, gb|N38122
come from this gene. [Arabidopsis thaliana]
gi|12083324|gb|AAG48821.1|AF332458_1 putative class I
chitinase [Arabidopsis thaliana]
gi|14334488|gb|AAK59442.1| putative class I chitinase
[Arabidopsis thaliana]
gi|17226329|gb|AAL37736.1|AF422178_1 chitinase-like
protein 1 [Arabidopsis thaliana]
gi|17226331|gb|AAL37737.1|AF422179_1 chitinase-like
protein 1 [Arabidopsis thaliana]
gi|21280935|gb|AAM44973.1| putative class I chitinase
[Arabidopsis thaliana]
Length = 321
Score = 237 bits (605), Expect = 6e-62
Identities = 111/160 (69%), Positives = 123/160 (76%), Gaps = 3/160 (1%)
Frame = +2
Query: 56 LVIFLAVSS---TVEGDDNTKTLVKYKHGKKYCDKGWECKGWSIYCCNLTISDYFQVYQF 226
+++ LAVS G+D T VK G K C +GWEC WS YCCN TISDYFQVYQF
Sbjct: 11 ILVLLAVSFLALVANGEDKT-IKVKKVRGNKVCTQGWECSWWSKYCCNQTISDYFQVYQF 69
Query: 227 ENLFSKRNTPVANAVGFWDYHSFITAAALFEPQGFGTTGNKTLQMMEIAAFLGHVGAKTS 406
E LFSKRNTP+A+AVGFWDY SFITAAALFEP GFGTTG K + E+AAFLGHV +KTS
Sbjct: 70 EQLFSKRNTPIAHAVGFWDYQSFITAAALFEPLGFGTTGGKLMGQKEMAAFLGHVASKTS 129
Query: 407 CGYGVATGGPLAWGLCYNHEMSPSQKYCDEYYKLTYPCTP 526
CGYGVATGGPLAWGLCYN EMSP Q YCDE +K YPC+P
Sbjct: 130 CGYGVATGGPLAWGLCYNREMSPMQSYCDESWKFKYPCSP 169
>dbj|BAC55635.1| OJ1081_B12.16 [Oryza sativa (japonica cultivar-group)]
Length = 316
Score = 204 bits (520), Expect = 4e-52
Identities = 95/129 (73%), Positives = 107/129 (82%), Gaps = 1/129 (0%)
Frame = +2
Query: 137 KYCDKGWECKGWSIYCCNLTISDYFQVYQFENLFSKRNT-PVANAVGFWDYHSFITAAAL 313
K CDKGWEC G S +CCN TI+DYF+ YQFE LF+ RN +A+A GFWDYH+FITAAAL
Sbjct: 40 KACDKGWECSG-SRFCCNDTITDYFKAYQFEELFAHRNDRSLAHAAGFWDYHAFITAAAL 98
Query: 314 FEPQGFGTTGNKTLQMMEIAAFLGHVGAKTSCGYGVATGGPLAWGLCYNHEMSPSQKYCD 493
FEP+GFGTTG K + M E+AAFLGHVGAKTSCGY VATGGPLAWGLCYNHE+SPSQ YCD
Sbjct: 99 FEPRGFGTTGGKEVGMKEVAAFLGHVGAKTSCGYSVATGGPLAWGLCYNHELSPSQSYCD 158
Query: 494 EYYKLTYPC 520
+L YPC
Sbjct: 159 NSNEL-YPC 166
>gb|AAF69789.1|AF135149_1 class I chitinase [Arabis microphylla]
Length = 299
Score = 92.4 bits (228), Expect = 3e-18
Identities = 45/110 (40%), Positives = 63/110 (56%)
Frame = +2
Query: 197 ISDYFQVYQFENLFSKRNTPVANAVGFWDYHSFITAAALFEPQGFGTTGNKTLQMMEIAA 376
+SD QF+++ RN A GF+ Y++FITAA F GFGTTG+ T + EIAA
Sbjct: 51 LSDIISSSQFDDMLMHRNDAACPARGFYTYNAFITAAKSFP--GFGTTGDTTTRKKEIAA 108
Query: 377 FLGHVGAKTSCGYGVATGGPLAWGLCYNHEMSPSQKYCDEYYKLTYPCTP 526
F G +T+ G+ A GP +WG C+ E +P+ YC+ T+PC P
Sbjct: 109 FFGQTSHETTGGWASAPDGPFSWGYCFKQEQNPTSNYCEP--SATWPCAP 156
>gb|AAF69774.1|AF135134_1 class I chitinase [Arabis blepharophylla]
Length = 289
Score = 90.5 bits (223), Expect = 1e-17
Identities = 47/124 (37%), Positives = 65/124 (51%), Gaps = 2/124 (1%)
Frame = +2
Query: 161 CK--GWSIYCCNLTISDYFQVYQFENLFSKRNTPVANAVGFWDYHSFITAAALFEPQGFG 334
CK G C +SD QF+++ RN A GF+ Y++FITAA F GFG
Sbjct: 27 CKLPGCQSQCTPGDLSDIISSSQFDDMLKNRNAASCPARGFYTYNAFITAAKSFP--GFG 84
Query: 335 TTGNKTLQMMEIAAFLGHVGAKTSCGYGVATGGPLAWGLCYNHEMSPSQKYCDEYYKLTY 514
TTG+ + E+AAF G +T+ G+ A GP +WG C+ E +P+ YC T+
Sbjct: 85 TTGDTATRKKEVAAFFGQTSHETTGGWASAPDGPFSWGYCFKQEQNPTSDYCQP--SATW 142
Query: 515 PCTP 526
PC P
Sbjct: 143 PCAP 146
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 536,367,346
Number of Sequences: 1393205
Number of extensions: 13192200
Number of successful extensions: 50659
Number of sequences better than 10.0: 383
Number of HSP's better than 10.0 without gapping: 46161
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 50208
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)