Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
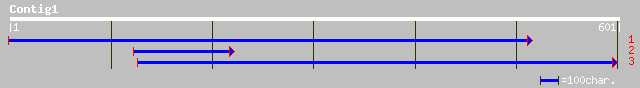
Query= KMC005129A_C01 KMC005129A_c01
(601 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_201056.1| microtubule-associated protein EB1-like protein... 77 3e-14
ref|NP_190353.1| putative protein; protein id: At3g47690.1 [Arab... 72 2e-13
gb|AAM93680.1| putative microtubule-associated protein [Oryza sa... 62 6e-09
ref|NP_201528.1| putative protein; protein id: At5g67270.1, supp... 60 2e-08
gb|AAA96321.1| APC-binding protein EB2 59 6e-08
>ref|NP_201056.1| microtubule-associated protein EB1-like protein; protein id:
At5g62500.1, supported by cDNA: 112876. [Arabidopsis
thaliana] gi|10178081|dbj|BAB11500.1|
microtubule-associated protein EB1-like protein
[Arabidopsis thaliana] gi|21536831|gb|AAM61163.1|
microtubule-associated protein EB1-like protein
[Arabidopsis thaliana]
Length = 293
Score = 77.0 bits (188), Expect(2) = 3e-14
Identities = 39/52 (75%), Positives = 44/52 (84%)
Frame = -2
Query: 600 LSVDLLEKERDFYFSNLRDIEILCPTTELENEPMSVAVTKILYAADASESPL 445
+SVDLLEKERDFYFS LRDIEILC T EL++ P+ VAV KILYA DA+ES L
Sbjct: 200 VSVDLLEKERDFYFSKLRDIEILCQTPELDDLPIVVAVKKILYATDANESVL 251
Score = 22.7 bits (47), Expect(2) = 3e-14
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 2/28 (7%)
Frame = -1
Query: 451 TITTEAQEYLNQT--LHAVEEEPEPEPE 374
++ EAQE LNQ+ L EEE + E E
Sbjct: 249 SVLEEAQECLNQSLGLEGYEEEGKEEEE 276
>ref|NP_190353.1| putative protein; protein id: At3g47690.1 [Arabidopsis thaliana]
gi|7487371|pir||T07708 hypothetical protein T23J7.20 -
Arabidopsis thaliana gi|4741186|emb|CAB41852.1| putative
protein [Arabidopsis thaliana]
Length = 155
Score = 72.0 bits (175), Expect(2) = 2e-13
Identities = 38/59 (64%), Positives = 44/59 (74%), Gaps = 7/59 (11%)
Frame = -2
Query: 600 LSVDLLEKERDFYFSNLRDIEILCPTTELEN-------EPMSVAVTKILYAADASESPL 445
+S DLLEKERDFYFS LRD+EILC T EL++ EP+ VAV KILYA DA+ES L
Sbjct: 65 ISTDLLEKERDFYFSKLRDVEILCQTPELDDLPVNVSVEPIVVAVKKILYATDANESAL 123
Score = 25.0 bits (53), Expect(2) = 2e-13
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = -1
Query: 439 EAQEYLNQTLHAVEEEPEPEPE 374
+AQEYLNQ+L ++E E E
Sbjct: 125 DAQEYLNQSLGVEDDEAEGNGE 146
>gb|AAM93680.1| putative microtubule-associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 332
Score = 62.0 bits (149), Expect = 6e-09
Identities = 30/50 (60%), Positives = 38/50 (76%)
Frame = -2
Query: 600 LSVDLLEKERDFYFSNLRDIEILCPTTELENEPMSVAVTKILYAADASES 451
L VD LEKERDFYFS LRD+EILC + E+E+ P+ A+ K+LYAA+ S
Sbjct: 206 LLVDSLEKERDFYFSKLRDVEILCQSPEVEHLPIVNAIHKVLYAAEDDPS 255
>ref|NP_201528.1| putative protein; protein id: At5g67270.1, supported by cDNA:
38358. [Arabidopsis thaliana] gi|9759276|dbj|BAB09646.1|
contains similarity to APC-binding protein
EB1~gene_id:K3G17.3 [Arabidopsis thaliana]
gi|21593362|gb|AAM65311.1| microtubule-associated
protein EB1-like protein [Arabidopsis thaliana]
Length = 329
Score = 60.1 bits (144), Expect = 2e-08
Identities = 27/49 (55%), Positives = 36/49 (73%)
Frame = -2
Query: 600 LSVDLLEKERDFYFSNLRDIEILCPTTELENEPMSVAVTKILYAADASE 454
L +D LEKERDFYFS LRD+EILC + E+ P+ ++ +ILYAAD +
Sbjct: 213 LYIDSLEKERDFYFSKLRDVEILCQNPDTEHLPLVGSIKRILYAADGED 261
>gb|AAA96321.1| APC-binding protein EB2
Length = 225
Score = 58.5 bits (140), Expect = 6e-08
Identities = 28/51 (54%), Positives = 35/51 (67%)
Frame = -2
Query: 600 LSVDLLEKERDFYFSNLRDIEILCPTTELENEPMSVAVTKILYAADASESP 448
L+VD LEKERDFYFS LRDIE++C E EN P+ + ILYA + +P
Sbjct: 158 LTVDGLEKERDFYFSKLRDIELICQEHESENSPVISGIIGILYATEEGFAP 208
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 507,770,988
Number of Sequences: 1393205
Number of extensions: 10754500
Number of successful extensions: 31633
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 26697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31158
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)