Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
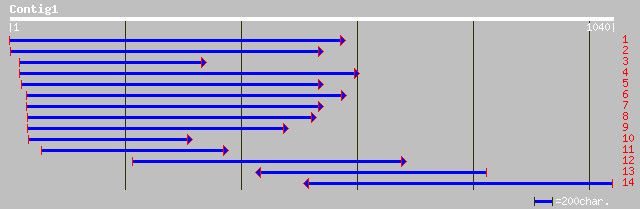
Query= KMC005127A_C01 KMC005127A_c01
(1016 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_084801.1| ATP synthase CF0 C chain [Lotus japonicus] gi|1... 153 5e-36
ref|NP_054483.1| ATP synthase CF0 C chain [Nicotiana tabacum] gi... 152 8e-36
ref|NP_039280.1| ATP synthase CF0 C chain [Marchantia polymorpha... 152 1e-35
ref|NP_051046.1| ATP synthase CF0 C chain [Arabidopsis thaliana]... 152 1e-35
ref|NP_039378.1| ATP synthase CF0 C chain [Oryza sativa (japonic... 151 2e-35
>ref|NP_084801.1| ATP synthase CF0 C chain [Lotus japonicus]
gi|114652|sp|P12231|ATPH_LOTJA ATP synthase C chain
(Lipid-binding protein) (Subunit III)
gi|81820|pir||A29599 H+-transporting two-sector ATPase
(EC 3.6.3.14) lipid-binding protein - soybean
chloroplast gi|169935|gb|AAB01580.1| DCCD-binding
protein gi|13358982|dbj|BAB33199.1| ATPase III subunit
[Lotus japonicus]
Length = 81
Score = 153 bits (386), Expect = 5e-36
Identities = 81/81 (100%), Positives = 81/81 (100%)
Frame = -3
Query: 762 MNPIISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 583
MNPIISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM
Sbjct: 1 MNPIISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 60
Query: 582 EALTIYGLVVALALLFANPFV 520
EALTIYGLVVALALLFANPFV
Sbjct: 61 EALTIYGLVVALALLFANPFV 81
>ref|NP_054483.1| ATP synthase CF0 C chain [Nicotiana tabacum]
gi|18860298|ref|NP_569616.1| ATP synthase CF0 C chain
[Psilotum nudum] gi|28202154|ref|NP_777395.1| ATPase III
subunit [Anthoceros formosae]
gi|114654|sp|P06286|ATPH_TOBAC ATP synthase C chain
(Lipid-binding protein) (Subunit III)
gi|67898|pir||LWNTA H+-transporting two-sector ATPase
(EC 3.6.3.14) lipid-binding protein - common tobacco
chloroplast gi|11812|emb|CAA77343.1| ATPase III subunit
[Nicotiana tabacum] gi|343484|gb|AAA84678.1| ATPase
subunit III gi|18389451|dbj|BAB84203.1| ATP synthase
subunit CF0 III [Psilotum nudum]
gi|27807856|dbj|BAC55331.1| ATPase III subunit
[Anthoceros formosae] gi|27807955|dbj|BAC55422.1| ATPase
III subunit [Anthoceros formosae]
gi|224347|prf||1102209A ATPase III,H translocating
gi|225272|prf||1211235G ATPase III
Length = 81
Score = 152 bits (384), Expect = 8e-36
Identities = 80/81 (98%), Positives = 81/81 (99%)
Frame = -3
Query: 762 MNPIISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 583
MNP+ISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM
Sbjct: 1 MNPLISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 60
Query: 582 EALTIYGLVVALALLFANPFV 520
EALTIYGLVVALALLFANPFV
Sbjct: 61 EALTIYGLVVALALLFANPFV 81
>ref|NP_039280.1| ATP synthase CF0 C chain [Marchantia polymorpha]
gi|13518334|ref|NP_084693.1| ATP synthase CF0 C chain
[Oenothera elata subsp. hookeri]
gi|114649|sp|P06287|ATPH_MARPO ATP synthase C chain
(Lipid-binding protein) (Subunit III)
gi|67899|pir||LWLVA H+-transporting two-sector ATPase
(EC 3.6.3.14) lipid-binding protein - liverwort
(Marchantia polymorpha) chloroplast
gi|11653|emb|CAA28066.1| atpH [Marchantia polymorpha]
gi|6723749|emb|CAB67158.1| ATP synthase subunit III
[Oenothera elata subsp. hookeri]
Length = 81
Score = 152 bits (383), Expect = 1e-35
Identities = 79/81 (97%), Positives = 81/81 (99%)
Frame = -3
Query: 762 MNPIISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 583
MNP+ISAASVIAAGLAVGLASIGPG+GQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM
Sbjct: 1 MNPLISAASVIAAGLAVGLASIGPGIGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 60
Query: 582 EALTIYGLVVALALLFANPFV 520
EALTIYGLVVALALLFANPFV
Sbjct: 61 EALTIYGLVVALALLFANPFV 81
>ref|NP_051046.1| ATP synthase CF0 C chain [Arabidopsis thaliana]
gi|6685247|sp|P56760|ATPH_ARATH ATP synthase C chain
(Lipid-binding protein) (Subunit III)
gi|5881681|dbj|BAA84372.1| ATPase III subunit
[Arabidopsis thaliana]
Length = 81
Score = 152 bits (383), Expect = 1e-35
Identities = 79/81 (97%), Positives = 81/81 (99%)
Frame = -3
Query: 762 MNPIISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 583
MNP++SAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM
Sbjct: 1 MNPLVSAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 60
Query: 582 EALTIYGLVVALALLFANPFV 520
EALTIYGLVVALALLFANPFV
Sbjct: 61 EALTIYGLVVALALLFANPFV 81
>ref|NP_039378.1| ATP synthase CF0 C chain [Oryza sativa (japonica cultivar-group)]
gi|11467187|ref|NP_043020.1| ATP synthase CF0 C chain
[Zea mays] gi|11497511|ref|NP_054919.1| ATP synthase CF0
C chain [Spinacia oleracea] gi|14017567|ref|NP_114254.1|
ATP synthase CF0 C chain [Triticum aestivum]
gi|114653|sp|P00843|ATPH_SPIOL ATP synthase C chain
(Lipid-binding protein) (Subunit III)
gi|67896|pir||LWSPA H+-transporting two-sector ATPase
(EC 3.6.3.14) lipid-binding protein - spinach
chloroplast gi|67897|pir||LWRZA H+-transporting
two-sector ATPase (EC 3.6.3.14) lipid-binding protein -
rice chloroplast gi|279576|pir||LWZMC H+-transporting
two-sector ATPase (EC 3.6.3.14) lipid-binding protein -
maize chloroplast gi|11976|emb|CAA33991.1| ATPase III
subunit [Oryza sativa (japonica cultivar-group)]
gi|12251|emb|CAA27402.1| CFo subunit III (atpH) (aa
1-81) proteolipid [Spinacia oleracea]
gi|12407|emb|CAA28875.1| CF(0) subunit III (AA 1-81)
[Zea mays] gi|342580|gb|AAA84473.1| phosphorylation
coupling factor alpha subunit (atpA)
gi|343673|gb|AAA84724.1| ATP synthase
proton-translocating subunit gi|902217|emb|CAA60281.1|
ATPase subunit III [Zea mays] gi|7636092|emb|CAB88712.1|
ATPase subunit III [Spinacia oleracea]
gi|13928200|dbj|BAB47029.1| ATPase III subunit [Triticum
aestivum] gi|19920163|gb|AAM08595.1|AC092750_29 Putative
ATPase III subunit from chromosome 10 chloroplast
insertion [Oryza sativa (japonica cultivar-group)]
gi|20143585|gb|AAM12342.1|AC091680_43 ATPase III subunit
[Oryza sativa (japonica cultivar-group)]
gi|20146748|gb|AAM12484.1|AC074232_11 ATPase III subunit
[Oryza sativa (japonica cultivar-group)]
gi|21327355|gb|AAM48260.1|AC122148_13 Putative ATPase
III subunit from chromosome 10 chloroplast insertion
[Oryza sativa (japonica cultivar-group)]
gi|223270|prf||0702201A synthase proteolipid subunit,ATP
gi|226694|prf||1603356V ATPase III
Length = 81
Score = 151 bits (381), Expect = 2e-35
Identities = 79/81 (97%), Positives = 81/81 (99%)
Frame = -3
Query: 762 MNPIISAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 583
MNP+I+AASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM
Sbjct: 1 MNPLIAAASVIAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFM 60
Query: 582 EALTIYGLVVALALLFANPFV 520
EALTIYGLVVALALLFANPFV
Sbjct: 61 EALTIYGLVVALALLFANPFV 81
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 822,580,274
Number of Sequences: 1393205
Number of extensions: 18648453
Number of successful extensions: 55947
Number of sequences better than 10.0: 424
Number of HSP's better than 10.0 without gapping: 49353
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 55417
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 59049410978
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)