Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
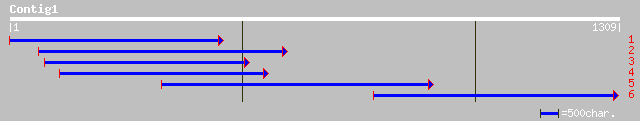
Query= KMC005120A_C01 KMC005120A_c01
(1306 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF20931.1|AF206721_1 ascorbate oxidase [Brassica juncea] 212 3e-87
emb|CAA75577.1| L-ascorbate oxidase [Medicago truncatula] 279 6e-74
ref|NP_197609.1| L-ascorbate oxidase, putative; protein id: At5g... 206 6e-73
gb|AAF35911.2|AF233594_1 ascorbate oxidase AO4 [Cucumis melo] 226 5e-58
sp|P14133|ASO_CUCSA L-ASCORBATE OXIDASE PRECURSOR (ASCORBASE) (A... 226 8e-58
>gb|AAF20931.1|AF206721_1 ascorbate oxidase [Brassica juncea]
Length = 574
Score = 212 bits (539), Expect(3) = 3e-87
Identities = 122/245 (49%), Positives = 149/245 (60%), Gaps = 13/245 (5%)
Frame = +2
Query: 395 PWADGTAAISQCAINPGETFHYWFTVDRPGTYFYHGHYGMQRAAGLYGSLIVDLPKGQNE 574
PWADG A ++QC INPGET Y F VD+ GT+FYHGHYGMQR+AGLYG +IV PK E
Sbjct: 91 PWADGAAGVTQCPINPGETLTYNFIVDKAGTHFYHGHYGMQRSAGLYGMMIVRSPK---E 147
Query: 575 PFHYDGEFNLLLSDLWHKSSHEQEVGLSSKPLKWIGEPQSLLINGRGQFNCSLAAKFVNT 754
YDGEFNLLLSD WH+SSH QE+ LSS+P++WIGEPQSLLINGRGQFNCSLAA F N
Sbjct: 148 TLQYDGEFNLLLSDWWHQSSHAQELYLSSRPMRWIGEPQSLLINGRGQFNCSLAAYF-NK 206
Query: 755 SLVPQCQFKGGEECA-------------ARDSSCGAKQELQDQNC*YHFHGFSQLGHFKS 895
+ +C+FK +ECA R +S A L F+ GH
Sbjct: 207 GGLKECKFKENDECAPSILRVEPNKVYRLRIASTTALASL----------NFAVEGHQLE 256
Query: 896 QADSGGSRRQPRPTLRRRRHRHLLRRDLLRSSSPPTRTQTTGYGSSVGVRGRKPATPQAL 1075
++ G+ P + LLR+ +P T + Y SVGVRGRKP T QAL
Sbjct: 257 VVEADGNYIAPFTVDDIDLYSGETYSVLLRTHNP---TPSRNYWISVGVRGRKPNTTQAL 313
Query: 1076 TIVNY 1090
T+++Y
Sbjct: 314 TVLHY 318
Score = 100 bits (250), Expect = 4e-20
Identities = 62/148 (41%), Positives = 76/148 (50%), Gaps = 3/148 (2%)
Frame = +3
Query: 801 PEILHVEPNKSYRIRIASTTSMASLNLAISNHKLIVVEADGNHVQPFAVDDIDIYSGETY 980
P IL VEPNK YR+RIASTT++ASLN A+ H+L VVEADGN++ PF VDDID+YSGETY
Sbjct: 222 PSILRVEPNKVYRLRIASTTALASLNFAVEGHQLEVVEADGNYIAPFTVDDIDLYSGETY 281
Query: 981 SGPPH-HRPEPKPQGTARQSESEAENPPPHKPSPSSTTKQSPAPSSPRT--PPPPTPRMG 1151
S H P P P+ + + AP PPP TPR
Sbjct: 282 SVLLRTHNPTPSRNYWISVG---VRGRKPNTTQALTVLHYAGAPEFEHLPHPPPVTPRWD 338
Query: 1152 RFRTQLSLHQENHRGNGHPTTDINLPPP 1235
+ S + G+P PPP
Sbjct: 339 DYDRSKSFTNKIFAAKGYP------PPP 360
Score = 87.8 bits (216), Expect(3) = 3e-87
Identities = 41/69 (59%), Positives = 49/69 (70%)
Frame = +1
Query: 187 AKVRHYKFDVEYMFKKPDCLEHVVMGINGQFPGPTIKAEVGDTLDIALTNKLHTEGTVIH 366
A V ++VEY PDC E +V+ ING+FPGPTI A GDT+ + +TNKL TEG VIH
Sbjct: 22 AGVVELNWEVEYKLGWPDCKEGIVIAINGEFPGPTINAVAGDTVIVHVTNKLSTEGVVIH 81
Query: 367 WHGIRQLGT 393
WHGIRQ GT
Sbjct: 82 WHGIRQNGT 90
Score = 67.8 bits (164), Expect(3) = 3e-87
Identities = 34/64 (53%), Positives = 41/64 (63%), Gaps = 3/64 (4%)
Frame = +1
Query: 1123 HRHHPPR---EWDDFERS*AFTKKITVGMGTPQPT*ISHRQIHLLNTQNLCDGFSEWAIN 1293
H HPP WDD++RS +FT KI G P P S Q+ LLNTQNL D +++WAIN
Sbjct: 326 HLPHPPPVTPRWDDYDRSKSFTNKIFAAKGYPPPPEKSDEQLFLLNTQNLMDKYTKWAIN 385
Query: 1294 NVSL 1305
NVSL
Sbjct: 386 NVSL 389
>emb|CAA75577.1| L-ascorbate oxidase [Medicago truncatula]
Length = 569
Score = 279 bits (714), Expect = 6e-74
Identities = 157/279 (56%), Positives = 179/279 (63%), Gaps = 14/279 (5%)
Frame = +2
Query: 395 PWADGTAAISQCAINPGETFHYWFTVDRPGTYFYHGHYGMQRAAGLYGSLIVDLPKGQNE 574
PWADGTAAISQCAINPGETF Y F VDRPGTYFYHGHYGMQRAAGLYGSLIVDLPK Q E
Sbjct: 92 PWADGTAAISQCAINPGETFQYKFKVDRPGTYFYHGHYGMQRAAGLYGSLIVDLPKSQRE 151
Query: 575 PFHYDGEFNLLLSDLWHKSSHEQEVGLSSKPLKWIGEPQSLLINGRGQFNCSLAAKFVNT 754
PFHYDGEFNLLLSDLWH SSHEQEVGLSS P++WIGEPQ+LLINGR QF+CSLA+K+ +T
Sbjct: 152 PFHYDGEFNLLLSDLWHTSSHEQEVGLSSAPMRWIGEPQNLLINGRRQFHCSLASKYGST 211
Query: 755 SLVPQCQFKGGEECAARDSSCGAKQELQDQNC*YHFHGFSQL---GHFKSQADSGGSRRQ 925
+L PQC KGGEECA + K+ + + L H ++ G+
Sbjct: 212 NL-PQCNLKGGEECAPQILHVEPKKTYRIRIASTTSLASLNLAISNHKLIVVEADGNYVH 270
Query: 926 PRPTLRRRRHRHLLRRDLLRSSSPPTRTQTTGYGSSVGVRGRKPATPQALTIVNYKTISX 1105
P + LL + P + Y S+GVRGRKP+TPQALTI+NYK +S
Sbjct: 271 PFAVDDIDIYSGETYSVLLTTDQDPKK----NYWLSIGVRGRKPSTPQALTILNYKPLSA 326
Query: 1106 SVFPTHTATTHPA-NGT----------ISNAAKPSPRKS 1189
SVFPT P N T IS P P KS
Sbjct: 327 SVFPTSPPPVTPLWNDTDHSKAFTKQIISKMGNPQPPKS 365
Score = 152 bits (384), Expect = 1e-35
Identities = 75/114 (65%), Positives = 84/114 (72%)
Frame = +1
Query: 112 MSLLKALFLWCLLWVGLFQSSVVVGAKVRHYKFDVEYMFKKPDCLEHVVMGINGQFPGPT 291
MSLLKA+F+WC+ +GLF+ S+ G HYKFDVEY++KKPDC EHVVMGINGQFPGPT
Sbjct: 1 MSLLKAVFVWCIC-LGLFELSLAKGKS--HYKFDVEYIYKKPDCKEHVVMGINGQFPGPT 57
Query: 292 IKAEVGDTLDIALTNKLHTEGTVIHWHGIRQLGTALGRWNCCYLTVCYKPGRDF 453
I+AEVGDTL I LTNKLHTEGTVIHWHGIRQ GT PG F
Sbjct: 58 IRAEVGDTLVIDLTNKLHTEGTVIHWHGIRQFGTPWADGTAAISQCAINPGETF 111
Score = 118 bits (295), Expect = 2e-25
Identities = 67/116 (57%), Positives = 75/116 (63%), Gaps = 2/116 (1%)
Frame = +3
Query: 801 PEILHVEPNKSYRIRIASTTSMASLNLAISNHKLIVVEADGNHVQPFAVDDIDIYSGETY 980
P+ILHVEP K+YRIRIASTTS+ASLNLAISNHKLIVVEADGN+V PFAVDDIDIYSGETY
Sbjct: 226 PQILHVEPKKTYRIRIASTTSLASLNLAISNHKLIVVEADGNYVHPFAVDDIDIYSGETY 285
Query: 981 SGPPHHRPEPKPQGTARQSESEAENPPPHKPSPSSTTKQSPAPSS--PRTPPPPTP 1142
S +PK P P + P +S P +PPP TP
Sbjct: 286 SVLLTTDQDPKKN---YWLSIGVRGRKPSTPQALTILNYKPLSASVFPTSPPPVTP 338
Score = 77.0 bits (188), Expect = 6e-13
Identities = 49/109 (44%), Positives = 59/109 (53%), Gaps = 2/109 (1%)
Frame = +1
Query: 985 VLLTTDQNPNHRVRLVSRSQRQKTRHPTSPHHRQLQNNLXLRLPHAHRHHPPRE--WDDF 1158
VLLTTDQ+P L S + R P++P + N L PP W+D
Sbjct: 287 VLLTTDQDPKKNYWL---SIGVRGRKPSTPQALTILNYKPLSASVFPTSPPPVTPLWNDT 343
Query: 1159 ERS*AFTKKITVGMGTPQPT*ISHRQIHLLNTQNLCDGFSEWAINNVSL 1305
+ S AFTK+I MG PQP SHR IH LNTQN F++WAINNVSL
Sbjct: 344 DHSKAFTKQIISKMGNPQPPKSSHRSIHFLNTQNKIGSFTKWAINNVSL 392
>ref|NP_197609.1| L-ascorbate oxidase, putative; protein id: At5g21100.1, supported by
cDNA: gi_20466240 [Arabidopsis thaliana]
gi|20466241|gb|AAM20438.1| ascorbate oxidase-like protein
[Arabidopsis thaliana] gi|28059566|gb|AAO30070.1|
ascorbate oxidase-like protein [Arabidopsis thaliana]
gi|29294063|gb|AAO73900.1| L-ascorbate oxidase, putative
[Arabidopsis thaliana]
Length = 573
Score = 206 bits (524), Expect(2) = 6e-73
Identities = 121/262 (46%), Positives = 151/262 (57%), Gaps = 13/262 (4%)
Frame = +2
Query: 395 PWADGTAAISQCAINPGETFHYWFTVDRPGTYFYHGHYGMQRAAGLYGSLIVDLPKGQNE 574
PWADG A ++QC INPGETF Y F VD+ GT+FYHGHYGMQR++GLYG LIV PK E
Sbjct: 90 PWADGAAGVTQCPINPGETFTYKFIVDKAGTHFYHGHYGMQRSSGLYGMLIVRSPK---E 146
Query: 575 PFHYDGEFNLLLSDLWHKSSHEQEVGLSSKPLKWIGEPQSLLINGRGQFNCSLAAKFVNT 754
YDGEFNLLLSD WH+S H QE+ LSS+P++WIGEPQSLLINGRGQFNCS AA F
Sbjct: 147 RLIYDGEFNLLLSDWWHQSIHAQELALSSRPMRWIGEPQSLLINGRGQFNCSQAAYFNKG 206
Query: 755 SLVPQCQFKGGEECA-------------ARDSSCGAKQELQDQNC*YHFHGFSQLGHFKS 895
C FK ++CA R +S A L + GH
Sbjct: 207 GEKDVCTFKENDQCAPQTLRVEPNRVYRLRIASTTALASL----------NLAVQGHQLV 256
Query: 896 QADSGGSRRQPRPTLRRRRHRHLLRRDLLRSSSPPTRTQTTGYGSSVGVRGRKPATPQAL 1075
++ G+ P + LL++++ P++ Y SVGVRGR+P TPQAL
Sbjct: 257 VVEADGNYVAPFTVNDIDVYSGETYSVLLKTNALPSKK----YWISVGVRGREPKTPQAL 312
Query: 1076 TIVNYKTISXSVFPTHTATTHP 1141
T++NY V T + +HP
Sbjct: 313 TVINY------VDATESRPSHP 328
Score = 97.8 bits (242), Expect = 3e-19
Identities = 58/137 (42%), Positives = 74/137 (53%), Gaps = 1/137 (0%)
Frame = +3
Query: 801 PEILHVEPNKSYRIRIASTTSMASLNLAISNHKLIVVEADGNHVQPFAVDDIDIYSGETY 980
P+ L VEPN+ YR+RIASTT++ASLNLA+ H+L+VVEADGN+V PF V+DID+YSGETY
Sbjct: 222 PQTLRVEPNRVYRLRIASTTALASLNLAVQGHQLVVVEADGNYVAPFTVNDIDVYSGETY 281
Query: 981 SGPPHHRPEPKPQGTARQSESEAENPPPHKPSPSSTTKQSPAPSS-PRTPPPPTPRMGRF 1157
S P + E P P + A S P PPP TP
Sbjct: 282 SVLLKTNALPSKKYWISVGVRGRE---PKTPQALTVINYVDATESRPSHPPPVTPIWNDT 338
Query: 1158 RTQLSLHQENHRGNGHP 1208
S ++ G+P
Sbjct: 339 DRSKSFSKKIFAAKGYP 355
Score = 92.0 bits (227), Expect(2) = 6e-73
Identities = 50/91 (54%), Positives = 60/91 (64%), Gaps = 2/91 (2%)
Frame = +1
Query: 127 ALFLWCLLWVGL--FQSSVVVGAKVRHYKFDVEYMFKKPDCLEHVVMGINGQFPGPTIKA 300
A+ +W LL V + F S+ A V ++VEY + PDC E +VM INGQFPGPTI A
Sbjct: 2 AVIVWWLLTVVVVAFHSA---SAAVVESTWEVEYKYWWPDCKEGIVMAINGQFPGPTIDA 58
Query: 301 EVGDTLDIALTNKLHTEGTVIHWHGIRQLGT 393
GDT+ I + NKL TEG VIHWHGIRQ GT
Sbjct: 59 VAGDTVIIHVVNKLSTEGVVIHWHGIRQKGT 89
Score = 63.2 bits (152), Expect = 9e-09
Identities = 41/110 (37%), Positives = 61/110 (55%), Gaps = 3/110 (2%)
Frame = +1
Query: 985 VLLTTDQNPNHRVRLVSRSQRQKTRHPTSPHHRQLQNNLXLRLPHAHRHHPPRE---WDD 1155
VLL T+ P+ + + S + R P +P + N + + HPP W+D
Sbjct: 283 VLLKTNALPSKKYWI---SVGVRGREPKTPQALTVINYVDAT--ESRPSHPPPVTPIWND 337
Query: 1156 FERS*AFTKKITVGMGTPQPT*ISHRQIHLLNTQNLCDGFSEWAINNVSL 1305
+RS +F+KKI G P+P SH Q+ LLNTQNL + +++W+INNVSL
Sbjct: 338 TDRSKSFSKKIFAAKGYPKPPEKSHDQLILLNTQNLYEDYTKWSINNVSL 387
>gb|AAF35911.2|AF233594_1 ascorbate oxidase AO4 [Cucumis melo]
Length = 587
Score = 226 bits (577), Expect = 5e-58
Identities = 127/254 (50%), Positives = 155/254 (61%), Gaps = 13/254 (5%)
Frame = +2
Query: 395 PWADGTAAISQCAINPGETFHYWFTVDRPGTYFYHGHYGMQRAAGLYGSLIVDLPKGQNE 574
PWADGTA+ISQCAINPGETF Y F VD+ GTYFYHGH GMQR+AGLYGSLIVD +G +E
Sbjct: 105 PWADGTASISQCAINPGETFTYRFVVDKAGTYFYHGHLGMQRSAGLYGSLIVDPQEGTSE 164
Query: 575 PFHYDGEFNLLLSDLWHKSSHEQEVGLSSKPLKWIGEPQSLLINGRGQFNCSLAAKFVNT 754
PFHYD E NLLLSD WH+S H+QEVGLSSKP++WIGEPQS+LINGRGQF+CS+AAK+ N
Sbjct: 165 PFHYDEEINLLLSDWWHQSVHKQEVGLSSKPMRWIGEPQSILINGRGQFDCSIAAKYKNG 224
Query: 755 SLVPQCQFKGGEECA-------------ARDSSCGAKQELQDQNC*YHFHGFSQLGHFKS 895
+ QC+ G E+CA R +S A L F+ H
Sbjct: 225 --LKQCELSGKEQCAPFILHVHPKKIYRIRIASTTALASL----------NFAIGNHKLL 272
Query: 896 QADSGGSRRQPRPTLRRRRHRHLLRRDLLRSSSPPTRTQTTGYGSSVGVRGRKPATPQAL 1075
++ G+ QP T + L+ + P+ Y S+GVR R P TP L
Sbjct: 273 VVEADGNYVQPFSTSDIDIYSGESYSVLITTDQNPSE----NYWVSIGVRARLPKTPPGL 328
Query: 1076 TIVNYKTISXSVFP 1117
T++NY S S P
Sbjct: 329 TVLNYLPNSVSKLP 342
Score = 116 bits (290), Expect = 9e-25
Identities = 52/89 (58%), Positives = 62/89 (69%)
Frame = +1
Query: 190 KVRHYKFDVEYMFKKPDCLEHVVMGINGQFPGPTIKAEVGDTLDIALTNKLHTEGTVIHW 369
K+RHYK++VEYMF PDC+E++VMGINGQFPGPTI+A GD + + LTNKLHTEG VIHW
Sbjct: 37 KIRHYKWEVEYMFWSPDCVENIVMGINGQFPGPTIRANAGDMVVVELTNKLHTEGVVIHW 96
Query: 370 HGIRQLGTALGRWNCCYLTVCYKPGRDFS 456
HGI Q GT PG F+
Sbjct: 97 HGILQRGTPWADGTASISQCAINPGETFT 125
Score = 99.0 bits (245), Expect(2) = 2e-35
Identities = 47/61 (77%), Positives = 53/61 (86%)
Frame = +3
Query: 801 PEILHVEPNKSYRIRIASTTSMASLNLAISNHKLIVVEADGNHVQPFAVDDIDIYSGETY 980
P ILHV P K YRIRIASTT++ASLN AI NHKL+VVEADGN+VQPF+ DIDIYSGE+Y
Sbjct: 238 PFILHVHPKKIYRIRIASTTALASLNFAIGNHKLLVVEADGNYVQPFSTSDIDIYSGESY 297
Query: 981 S 983
S
Sbjct: 298 S 298
Score = 74.3 bits (181), Expect(2) = 2e-35
Identities = 48/109 (44%), Positives = 68/109 (62%), Gaps = 2/109 (1%)
Frame = +1
Query: 985 VLLTTDQNP--NHRVRLVSRSQRQKTRHPTSPHHRQLQNNLXLRLPHAHRHHPPREWDDF 1158
VL+TTDQNP N+ V + R++ KT P L N++ +LP + P +W+DF
Sbjct: 299 VLITTDQNPSENYWVSIGVRARLPKTP-PGLTVLNYLPNSVS-KLPISPPPETP-DWEDF 355
Query: 1159 ERS*AFTKKITVGMGTPQPT*ISHRQIHLLNTQNLCDGFSEWAINNVSL 1305
+RS FT +I MG+P+P +R++ LLNTQN +GF +WAINNVSL
Sbjct: 356 DRSKNFTFRIFAAMGSPKPPVRYNRRLFLLNTQNRINGFMKWAINNVSL 404
>sp|P14133|ASO_CUCSA L-ASCORBATE OXIDASE PRECURSOR (ASCORBASE) (ASO) gi|66291|pir||KSKVAO
L-ascorbate oxidase (EC 1.10.3.3) precursor - cucumber
gi|167513|gb|AAA33119.1| ascorbate oxidase precursor (EC
1.10.3.3)
Length = 587
Score = 226 bits (575), Expect = 8e-58
Identities = 128/254 (50%), Positives = 156/254 (61%), Gaps = 13/254 (5%)
Frame = +2
Query: 395 PWADGTAAISQCAINPGETFHYWFTVDRPGTYFYHGHYGMQRAAGLYGSLIVDLPKGQNE 574
PWADGTA+ISQCAINPGETF Y F VD+ GTYFYHGH GMQR+AGLYGSLIVD P+G++E
Sbjct: 105 PWADGTASISQCAINPGETFTYRFVVDKAGTYFYHGHLGMQRSAGLYGSLIVDPPEGRSE 164
Query: 575 PFHYDGEFNLLLSDLWHKSSHEQEVGLSSKPLKWIGEPQSLLINGRGQFNCSLAAKFVNT 754
PFHYD E NLLLSD WH+S H+QEVGLSSKP++WIGEPQS+LING+GQF+CS+AAK+ N
Sbjct: 165 PFHYDEEINLLLSDWWHQSVHKQEVGLSSKPMRWIGEPQSILINGKGQFDCSIAAKY-NQ 223
Query: 755 SLVPQCQFKGGEECA-------------ARDSSCGAKQELQDQNC*YHFHGFSQLGHFKS 895
L QC+ G E+CA R +S A L F+ H
Sbjct: 224 GL-KQCELSGKEKCAPFILHVQPKKTYRIRIASTTALASL----------NFAIGNHELL 272
Query: 896 QADSGGSRRQPRPTLRRRRHRHLLRRDLLRSSSPPTRTQTTGYGSSVGVRGRKPATPQAL 1075
++ G+ QP T + L+ + P Y S+GVR R P TP L
Sbjct: 273 VVEADGNYVQPFVTSDIDIYSGESYSVLITTDQNPLE----NYWVSIGVRARLPKTPPGL 328
Query: 1076 TIVNYKTISXSVFP 1117
T++NY S S P
Sbjct: 329 TLLNYLPNSASKLP 342
Score = 116 bits (290), Expect = 9e-25
Identities = 58/120 (48%), Positives = 72/120 (59%)
Frame = +1
Query: 97 K*FYTMSLLKALFLWCLLWVGLFQSSVVVGAKVRHYKFDVEYMFKKPDCLEHVVMGINGQ 276
K F+ L L L + G+ S K++HYK+DVEYMF PDC+E++VMGING+
Sbjct: 7 KPFFPKPFLSFLVLSIIFGFGITLSEAGF-PKIKHYKWDVEYMFWSPDCVENIVMGINGE 65
Query: 277 FPGPTIKAEVGDTLDIALTNKLHTEGTVIHWHGIRQLGTALGRWNCCYLTVCYKPGRDFS 456
FPGPTI+A GD + + LTNKLHTEG VIHWHGI Q GT PG F+
Sbjct: 66 FPGPTIRANAGDIVVVELTNKLHTEGVVIHWHGILQRGTPWADGTASISQCAINPGETFT 125
Score = 101 bits (251), Expect = 3e-20
Identities = 75/208 (36%), Positives = 94/208 (45%), Gaps = 61/208 (29%)
Frame = +3
Query: 801 PEILHVEPNKSYRIRIASTTSMASLNLAISNHKLIVVEADGNHVQPFAVDDIDIYSGETY 980
P ILHV+P K+YRIRIASTT++ASLN AI NH+L+VVEADGN+VQPF DIDIYSGE+Y
Sbjct: 238 PFILHVQPKKTYRIRIASTTALASLNFAIGNHELLVVEADGNYVQPFVTSDIDIYSGESY 297
Query: 981 S-----------------GPPHHRPEPKPQGT-----ARQSESEAENPPPHKP------- 1073
S G P+ P T + +PPP P
Sbjct: 298 SVLITTDQNPLENYWVSIGVRARLPKTPPGLTLLNYLPNSASKLPISPPPETPHWEDFDR 357
Query: 1074 SPSSTTKQSPAPSSPRTPP------------------------------PPTPRMGRFRT 1163
S + T + A SP+ P PPTP + +
Sbjct: 358 SKNFTFRIFAAMGSPKPPVRYNRRLFLLNTQNRINGFMKWAINNVSLALPPTPYLAAMKM 417
Query: 1164 QL-SLHQENHRGNGHPTT-DINLPPPNP 1241
+L + +N P DIN PPPNP
Sbjct: 418 RLNTAFNQNPPPETFPLNYDINNPPPNP 445
Score = 73.9 bits (180), Expect = 5e-12
Identities = 49/112 (43%), Positives = 66/112 (58%), Gaps = 5/112 (4%)
Frame = +1
Query: 985 VLLTTDQNP--NHRVRLVSRSQRQKTRHPTSPHHRQLQN---NLXLRLPHAHRHHPPREW 1149
VL+TTDQNP N+ V + R+ R P +P L N N +LP + P W
Sbjct: 299 VLITTDQNPLENYWVSIGVRA-----RLPKTPPGLTLLNYLPNSASKLPISPPPETPH-W 352
Query: 1150 DDFERS*AFTKKITVGMGTPQPT*ISHRQIHLLNTQNLCDGFSEWAINNVSL 1305
+DF+RS FT +I MG+P+P +R++ LLNTQN +GF +WAINNVSL
Sbjct: 353 EDFDRSKNFTFRIFAAMGSPKPPVRYNRRLFLLNTQNRINGFMKWAINNVSL 404
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,318,090,771
Number of Sequences: 1393205
Number of extensions: 36007625
Number of successful extensions: 317797
Number of sequences better than 10.0: 3632
Number of HSP's better than 10.0 without gapping: 141341
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 242231
length of database: 448,689,247
effective HSP length: 126
effective length of database: 273,145,417
effective search space used: 84128788436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)