Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005104A_C02 KMC005104A_c02
(995 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
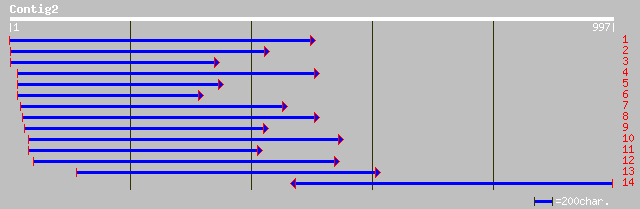
ref|NP_566741.1| expressed protein; protein id: At3g23840.1, sup... 133 5e-30
gb|AAM65391.1| fatty acid elongase-like protein (cer2-like) [Ara... 130 2e-29
ref|NP_194182.1| CER2; protein id: At4g24510.1, supported by cDN... 124 3e-27
pir||S60660 hypothetical protein - maize gi|949980|emb|CAA61258.... 122 1e-26
ref|NP_193120.1| fatty acid elongase - like protein (cer2-like);... 117 2e-25
>ref|NP_566741.1| expressed protein; protein id: At3g23840.1, supported by cDNA:
38849. [Arabidopsis thaliana] gi|9294653|dbj|BAB03002.1|
fatty acid elongase-like protein [Arabidopsis thaliana]
Length = 420
Score = 133 bits (334), Expect = 5e-30
Identities = 71/237 (29%), Positives = 128/237 (53%)
Frame = -2
Query: 964 SNLSEPQFPTKSLNIEKPSIIKKTRXVGEFWLATNDNDVATHTFHITSKQLQQLLTATNQ 785
++++E F + +KP +K+ VG+ W+A N++ + T +F++T L+
Sbjct: 191 TSVTERDFQNPTSTFKKPDSVKQVDLVGDLWVAPNNSKMTTFSFNLTVNDLKTHFPVNGD 250
Query: 784 TNNATKVEASYFEIISALVWKCIAQISDGSGPRVVTICTSDSNRAENEFPSNGLVLSKFE 605
FEI++ ++WKC+A + S P +T+ SD + + NG ++S
Sbjct: 251 GE---------FEILTGIIWKCVATVRGESAPVTITVIRSDPKKLKPRAVRNGQMISSIH 301
Query: 604 ASSAAGKSDVLKLAKLIAEKRTVENHAMEKLVDEGEGKEDFIVYGANLTFVDLEEADFRG 425
+ ++ + ++ K I E + E ++++VD+ DFIVYGANLTFVD+ E DF
Sbjct: 302 VDFSVAEASLEEIVKSIGEAKD-ERVVIDEIVDD---VSDFIVYGANLTFVDMSEVDFYE 357
Query: 424 VKLNGHKPVMANCTFRGVGDQGVVLVLPAPEDGGGDGRIVTVSLPGKELDQLKAKLE 254
K+ G P C +G+GD G V+VLP + + R+VTV+LP E++++K +++
Sbjct: 358 AKVMGKSPESVYCNVQGIGDDGAVVVLPGVVE---EERVVTVTLPVDEIEKVKWEMK 411
>gb|AAM65391.1| fatty acid elongase-like protein (cer2-like) [Arabidopsis thaliana]
Length = 420
Score = 130 bits (328), Expect = 2e-29
Identities = 70/230 (30%), Positives = 124/230 (53%)
Frame = -2
Query: 943 FPTKSLNIEKPSIIKKTRXVGEFWLATNDNDVATHTFHITSKQLQQLLTATNQTNNATKV 764
F + +++P IK+ VG+ W+A N++ + T +F++T L+
Sbjct: 198 FQNPTSTVKQPDSIKRVDPVGDLWVAPNNSKMTTFSFNLTVNDLKSNFPVNGDGE----- 252
Query: 763 EASYFEIISALVWKCIAQISDGSGPRVVTICTSDSNRAENEFPSNGLVLSKFEASSAAGK 584
FEI++ ++WKC+A + S P +T+ SD + + NG ++S + +
Sbjct: 253 ----FEILTGIIWKCVATVRGESAPVTITVIRSDPKKLKPRAVRNGQMISSIHVDFSVAE 308
Query: 583 SDVLKLAKLIAEKRTVENHAMEKLVDEGEGKEDFIVYGANLTFVDLEEADFRGVKLNGHK 404
+ + ++ K I E + E ++++VD+ DFIVYGANLTFVD+ E DF K+ G
Sbjct: 309 ASLEEIVKSIGEAKD-ERVVIDEIVDD---VSDFIVYGANLTFVDMSEVDFYEAKVMGKS 364
Query: 403 PVMANCTFRGVGDQGVVLVLPAPEDGGGDGRIVTVSLPGKELDQLKAKLE 254
P C +G+GD G V+VLP + + R+VTV+LP E++++K +++
Sbjct: 365 PESVYCNVQGIGDDGAVVVLPGVVE---EERVVTVTLPVDEIEKVKWEMK 411
>ref|NP_194182.1| CER2; protein id: At4g24510.1, supported by cDNA: 33382.
[Arabidopsis thaliana] gi|7459577|pir||T05583 CER2
protein - Arabidopsis thaliana
gi|1213594|emb|CAA63618.1| responsible for fatty acid
elongation from C28 to C30 [Arabidopsis thaliana]
gi|1655786|gb|AAB17946.1| CER2 gene product
gi|4220539|emb|CAA23012.1| CER2 [Arabidopsis thaliana]
gi|7269301|emb|CAB79361.1| CER2 [Arabidopsis thaliana]
gi|21592867|gb|AAM64817.1| CER2 [Arabidopsis thaliana]
Length = 421
Score = 124 bits (310), Expect = 3e-27
Identities = 81/247 (32%), Positives = 130/247 (51%), Gaps = 1/247 (0%)
Frame = -2
Query: 994 GNPPPKSLHISNLSEPQFPTKSLNIEKPSIIKKTRXVGEFWLATNDNDVATHTFHITSKQ 815
G+ P K ++ P+ + + N + I+K VGE+WL TN + H F+ +
Sbjct: 187 GHAPTKPVYPKT---PELTSHARNDGEAISIEKIDSVGEYWLLTNKCKMGRHIFNFSLNH 243
Query: 814 LQQLLTATNQTNNATKVEASYFEIISALVWKCIAQISDGSGPRVVTICT-SDSNRAENEF 638
+ L+ + S +I+ AL+WK + I + V+TIC S+ NE
Sbjct: 244 IDSLMAKYTTRDQPF----SEVDILYALIWKSLLNIRGETNTNVITICDRKKSSTCWNE- 298
Query: 637 PSNGLVLSKFEASSAAGKSDVLKLAKLIAEKRTVENHAMEKLVDEGEGKEDFIVYGANLT 458
LV+S E + + +LA LIA ++ EN A+++++++ +G DF YGANLT
Sbjct: 299 ---DLVISVVEKNDEM--VGISELAALIAGEKREENGAIKRMIEQDKGSSDFFTYGANLT 353
Query: 457 FVDLEEADFRGVKLNGHKPVMANCTFRGVGDQGVVLVLPAPEDGGGDGRIVTVSLPGKEL 278
FV+L+E D +++NG KP N T GVGD+GVVLV P RIV+V +P ++L
Sbjct: 354 FVNLDEIDMYELEINGGKPDFVNYTIHGVGDKGVVLVFPKQ----NFARIVSVVMPEEDL 409
Query: 277 DQLKAKL 257
+LK ++
Sbjct: 410 AKLKEEV 416
>pir||S60660 hypothetical protein - maize gi|949980|emb|CAA61258.1| orf [Zea
mays]
Length = 426
Score = 122 bits (305), Expect = 1e-26
Identities = 79/236 (33%), Positives = 124/236 (52%), Gaps = 2/236 (0%)
Frame = -2
Query: 958 LSEPQFPTKSLNIEKPSIIKKTRXVGEFWLATNDNDVATHTFHITSKQLQQLLTATNQTN 779
L+ P P + + P +K+ + + WL D+A ++FH++ L++L Q N
Sbjct: 198 LTPPNQPLQGQSPAAPRSVKQVGPIEDLWLVPAGRDMACYSFHVSDAVLKKL---HQQQN 254
Query: 778 NATKVEASYFEIISALVWKCIAQISDGSGPRVVTICTSDSNRAENEFPSNGLVLSKFE-A 602
A FE++SALVW+ +A+I VT+ +D+ + +N + + E A
Sbjct: 255 GRQDAAAGTFELVSALVWQAVAKIRGDVD--TVTVVRADAAGRSGKSLANEMKVGYVESA 312
Query: 601 SSAAGKSDVLKLAKLIAEKRTVENHAMEKLVDEGEGKEDFIVYG-ANLTFVDLEEADFRG 425
S+ K+D+ +LA L+A+ E A+ + D +VYG ANLT VD+E+ D G
Sbjct: 313 GSSPAKTDLAELAALLAKNLVDETAAVAAF------QGDVLVYGGANLTLVDMEQVDLYG 366
Query: 424 VKLNGHKPVMANCTFRGVGDQGVVLVLPAPEDGGGDGRIVTVSLPGKELDQLKAKL 257
+++ G +PV GVGD+G VLV P D G GR+VT LPG E+D L+A L
Sbjct: 367 LEIKGQRPVHVEYGMDGVGDEGAVLVQP---DADGRGRLVTAVLPGDEIDSLRAAL 419
>ref|NP_193120.1| fatty acid elongase - like protein (cer2-like); protein id:
At4g13840.1, supported by cDNA: gi_17933316 [Arabidopsis
thaliana] gi|7459578|pir||T05253 CER2 protein homolog
F18A5.230 - Arabidopsis thaliana
gi|4455313|emb|CAB36848.1| fatty acid elongase-like
protein (cer2-like) [Arabidopsis thaliana]
gi|7268088|emb|CAB78426.1| fatty acid elongase-like
protein (cer2-like) [Arabidopsis thaliana]
gi|17933317|gb|AAL48240.1|AF446369_1 AT4g13840/F18A5_230
[Arabidopsis thaliana] gi|23308387|gb|AAN18163.1|
At4g13840/F18A5_230 [Arabidopsis thaliana]
Length = 428
Score = 117 bits (294), Expect = 2e-25
Identities = 70/243 (28%), Positives = 128/243 (51%), Gaps = 1/243 (0%)
Frame = -2
Query: 961 NLSEPQFPTKSLNIEKPSIIKKTRXVGEFWLATNDNDVATHTFHIT-SKQLQQLLTATNQ 785
++ E +F + + ++ P IK+ VG+ W+ ND +A + F+++ + Q+ A
Sbjct: 195 SIGERRFQSPNPTVKDPVSIKRVEPVGDLWVTPNDKKLANYCFNLSVADQISPHFPAKGD 254
Query: 784 TNNATKVEASYFEIISALVWKCIAQISDGSGPRVVTICTSDSNRAENEFPSNGLVLSKFE 605
+ FEI++ ++WKCIA++ P VTI D N + N V+S
Sbjct: 255 DS------IPVFEILAGIIWKCIAKVRVEPKPVTVTIIKKDPNDLKLNAIRNSQVISSVS 308
Query: 604 ASSAAGKSDVLKLAKLIAEKRTVENHAMEKLVDEGEGKEDFIVYGANLTFVDLEEADFRG 425
++ V +L K + E + E +E++ + +G DF+VYGA LTF+DL D
Sbjct: 309 VDFPVAEATVEELVKAMGEAKD-ERCGIEEIGESCDGNLDFVVYGAKLTFLDLTGEDLYE 367
Query: 424 VKLNGHKPVMANCTFRGVGDQGVVLVLPAPEDGGGDGRIVTVSLPGKELDQLKAKLEGEW 245
K+ G P C G+G++G+V+V A + + R+VTV+LP +E++++K + + ++
Sbjct: 368 AKVMGKSPESVYCNVEGIGEEGLVVVYAAAK---SEERVVTVTLPEEEMERVKLEFK-KF 423
Query: 244 GII 236
G+I
Sbjct: 424 GLI 426
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 885,844,379
Number of Sequences: 1393205
Number of extensions: 21033468
Number of successful extensions: 84105
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 68340
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 81041
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 57117888189
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)