Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
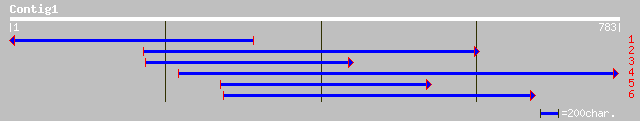
Query= KMC005095A_C01 KMC005095A_c01
(770 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565523.1| expressed protein; protein id: At2g21960.1, sup... 230 2e-59
gb|AAM64802.1| unknown [Arabidopsis thaliana] 228 7e-59
pir||C84607 hypothetical protein At2g21960 [imported] - Arabidop... 221 9e-57
ref|ZP_00071560.1| hypothetical protein [Trichodesmium erythraeu... 96 4e-19
gb|ZP_00107031.1| hypothetical protein [Nostoc punctiforme] 85 1e-15
>ref|NP_565523.1| expressed protein; protein id: At2g21960.1, supported by cDNA:
33232., supported by cDNA: gi_14334629 [Arabidopsis
thaliana] gi|14334630|gb|AAK59493.1| unknown protein
[Arabidopsis thaliana] gi|20198006|gb|AAD20413.2|
expressed protein [Arabidopsis thaliana]
gi|23296622|gb|AAN13134.1| unknown protein [Arabidopsis
thaliana]
Length = 332
Score = 230 bits (586), Expect = 2e-59
Identities = 111/142 (78%), Positives = 134/142 (94%)
Frame = -1
Query: 770 HEAAHFLIAYLLGVPILGYSLDIGKEHVNLIDQRLEKLIYSGQLNAKEIDRLAVVSMAGL 591
HEAAHFL+AYL+G+PILGYSLDIGKEHVNLID+RL KLIYSG+L++KE+DRLA V+MAGL
Sbjct: 191 HEAAHFLVAYLIGLPILGYSLDIGKEHVNLIDERLAKLIYSGKLDSKELDRLAAVAMAGL 250
Query: 590 AAEGLIYDKVIGQSADLFTLQRFINRTKPQLSKDQQQNLTRWAVMFAASLLKNNKESHEA 411
AAEGL YDKVIGQSADLF+LQRFINR++P++S +QQQNLTRWAV+++ASLLKNNK HEA
Sbjct: 251 AAEGLKYDKVIGQSADLFSLQRFINRSQPKISNEQQQNLTRWAVLYSASLLKNNKTIHEA 310
Query: 410 LMASMTKKASVVECIQTIESVA 345
LMA+M+K ASV+ECIQTIE+ +
Sbjct: 311 LMAAMSKNASVLECIQTIETAS 332
>gb|AAM64802.1| unknown [Arabidopsis thaliana]
Length = 332
Score = 228 bits (581), Expect = 7e-59
Identities = 110/142 (77%), Positives = 133/142 (93%)
Frame = -1
Query: 770 HEAAHFLIAYLLGVPILGYSLDIGKEHVNLIDQRLEKLIYSGQLNAKEIDRLAVVSMAGL 591
HEAAHFL+AYL+G+PILGYSLDIGKEHVNLID+RL KLIYSG+L++KE+DRLA V+MAGL
Sbjct: 191 HEAAHFLVAYLIGLPILGYSLDIGKEHVNLIDERLAKLIYSGKLDSKELDRLAAVAMAGL 250
Query: 590 AAEGLIYDKVIGQSADLFTLQRFINRTKPQLSKDQQQNLTRWAVMFAASLLKNNKESHEA 411
AAEGL YDKVIGQSADLF+LQRFINR++P++S +QQQNLTRWA +++ASLLKNNK HEA
Sbjct: 251 AAEGLKYDKVIGQSADLFSLQRFINRSQPKISNEQQQNLTRWAXLYSASLLKNNKTIHEA 310
Query: 410 LMASMTKKASVVECIQTIESVA 345
LMA+M+K ASV+ECIQTIE+ +
Sbjct: 311 LMAAMSKNASVLECIQTIETAS 332
>pir||C84607 hypothetical protein At2g21960 [imported] - Arabidopsis thaliana
Length = 344
Score = 221 bits (563), Expect = 9e-57
Identities = 111/154 (72%), Positives = 134/154 (86%), Gaps = 12/154 (7%)
Frame = -1
Query: 770 HEAAHFL------------IAYLLGVPILGYSLDIGKEHVNLIDQRLEKLIYSGQLNAKE 627
HEAAHFL +AYL+G+PILGYSLDIGKEHVNLID+RL KLIYSG+L++KE
Sbjct: 191 HEAAHFLGTLEKFKSFDSKVAYLIGLPILGYSLDIGKEHVNLIDERLAKLIYSGKLDSKE 250
Query: 626 IDRLAVVSMAGLAAEGLIYDKVIGQSADLFTLQRFINRTKPQLSKDQQQNLTRWAVMFAA 447
+DRLA V+MAGLAAEGL YDKVIGQSADLF+LQRFINR++P++S +QQQNLTRWAV+++A
Sbjct: 251 LDRLAAVAMAGLAAEGLKYDKVIGQSADLFSLQRFINRSQPKISNEQQQNLTRWAVLYSA 310
Query: 446 SLLKNNKESHEALMASMTKKASVVECIQTIESVA 345
SLLKNNK HEALMA+M+K ASV+ECIQTIE+ +
Sbjct: 311 SLLKNNKTIHEALMAAMSKNASVLECIQTIETAS 344
>ref|ZP_00071560.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 229
Score = 96.3 bits (238), Expect = 4e-19
Identities = 56/149 (37%), Positives = 90/149 (59%), Gaps = 10/149 (6%)
Frame = -1
Query: 770 HEAAHFLIAYLLGVPILGYSLDIGKEH---------VNLIDQRLEKLIYSGQLNAKEIDR 618
HEA HFL+AYLL +PI GY+L+ + V DQ+L +YSG ++++ +DR
Sbjct: 77 HEAGHFLVAYLLEIPISGYALNAWEAFRQGQSSQGGVRFDDQKLAAQLYSGVISSQLVDR 136
Query: 617 LAVVSMAGLAAEGLIYDKVIGQSADLFTLQRFINRTK-PQLSKDQQQNLTRWAVMFAASL 441
V MAG+AAE L+Y G + D + + + K P SK +Q WA + A +L
Sbjct: 137 YCTVWMAGIAAENLVYGNAEGGAEDRTKITAILRQLKRPGESKLKQS----WASLQARNL 192
Query: 440 LKNNKESHEALMASMTKKASVVECIQTIE 354
L+N++ +++AL+ +MT+++SV +C QTI+
Sbjct: 193 LENHQSAYKALVKAMTERSSVSDCYQTIK 221
>gb|ZP_00107031.1| hypothetical protein [Nostoc punctiforme]
Length = 225
Score = 85.1 bits (209), Expect = 1e-15
Identities = 53/143 (37%), Positives = 80/143 (55%), Gaps = 9/143 (6%)
Frame = -1
Query: 770 HEAAHFLIAYLLGVPILGYSLDI---------GKEHVNLIDQRLEKLIYSGQLNAKEIDR 618
HEA HFL+AYLLG+P+ GY+L G+ V+ D L + G+++A+ +DR
Sbjct: 77 HEAGHFLVAYLLGIPVTGYTLSAWEAWKQGQPGQGGVSFDDGELASQLEVGKISAQMLDR 136
Query: 617 LAVVSMAGLAAEGLIYDKVIGQSADLFTLQRFINRTKPQLSKDQQQNLTRWAVMFAASLL 438
V MAG+AAE L++D G S D L + T S+ Q R+ + A +LL
Sbjct: 137 YCTVWMAGIAAETLVFDNAEGGSDDKSKLIGVL--TVLGFSESVYQQKLRFHALQAKTLL 194
Query: 437 KNNKESHEALMASMTKKASVVEC 369
+ N S+EAL+ +M ++ASV +C
Sbjct: 195 QENWSSYEALVNAMRQRASVEDC 217
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 598,616,749
Number of Sequences: 1393205
Number of extensions: 11771007
Number of successful extensions: 26726
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 25992
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26689
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37815044670
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)