Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
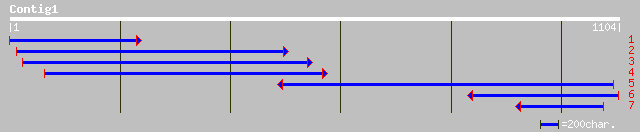
Query= KMC005081A_C01 KMC005081A_c01
(1096 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC33003.1| urease accessory protein G [Solanum tuberosum] 365 e-127
emb|CAC33000.1| urease accessory protein G [Solanum tuberosum] 366 e-127
ref|NP_180994.1| putative urease accessory protein; protein id: ... 369 e-126
emb|CAC33002.1| urease accessory protein G [Solanum tuberosum] 364 e-126
gb|AAD44338.1|AF160197_1 Ni-binding urease accessory protein Ure... 353 e-125
>emb|CAC33003.1| urease accessory protein G [Solanum tuberosum]
Length = 284
Score = 365 bits (938), Expect(2) = e-127
Identities = 182/208 (87%), Positives = 195/208 (93%)
Frame = -1
Query: 880 HFNERAFTIGRSAGPVGTGKTALMLALCEFLRDNYSLAAVTNDIFTKEDGEFLVRNKALP 701
+F+ERAFTIG GPVGTGKTALMLALC+ LR+ YSLAAVTNDIFTKEDGEFL+++ ALP
Sbjct: 77 NFSERAFTIG-IGGPVGTGKTALMLALCKVLREKYSLAAVTNDIFTKEDGEFLIKHGALP 135
Query: 700 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKTDILLCESGGDNLAANFSRELADY 521
EERIRAVETGGCPHAAIREDISINLGPLEELSNLYK DILLCESGGDNLAANFSRELADY
Sbjct: 136 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKADILLCESGGDNLAANFSRELADY 195
Query: 520 IIYIIDVSGGDKIPRKGGPGITQADLLVINKTDLAQAIGADLGVMERDSLRMRDGGPFVF 341
IIYIIDVS GDKIPRKGGPGITQADLLVINKTDLA A+GADL VMERD+LRMRDGGPFVF
Sbjct: 196 IIYIIDVSAGDKIPRKGGPGITQADLLVINKTDLAAAVGADLSVMERDALRMRDGGPFVF 255
Query: 340 AQVKHKVGVEEIVNHVIQAWEAATGKKR 257
AQVKH VGVE+IVNH++QAWE ATG K+
Sbjct: 256 AQVKHGVGVEDIVNHILQAWEVATGNKK 283
Score = 114 bits (286), Expect(2) = e-127
Identities = 49/78 (62%), Positives = 58/78 (73%)
Frame = -3
Query: 1094 HHHHHSDHGHDHDHHDNSHSHSDGVASAGSDSYVGPDGKVYHSHDGLAPHSHEPIYSPGF 915
HH H DH HDHDH + H+H + S+VG DGKVYHSHDGLAPHSHEPIYSPG+
Sbjct: 6 HHMHDHDHDHDHDHDHHHHNHDHTHEDTKATSWVGADGKVYHSHDGLAPHSHEPIYSPGY 65
Query: 914 FTRRAPPLINRAFQ*KSF 861
F+RRAPPLI+R F ++F
Sbjct: 66 FSRRAPPLIDRNFSERAF 83
>emb|CAC33000.1| urease accessory protein G [Solanum tuberosum]
Length = 278
Score = 366 bits (939), Expect(2) = e-127
Identities = 182/208 (87%), Positives = 195/208 (93%)
Frame = -1
Query: 880 HFNERAFTIGRSAGPVGTGKTALMLALCEFLRDNYSLAAVTNDIFTKEDGEFLVRNKALP 701
+F+ERAFTIG GPVGTGKTALMLALC+ LRD YSLAAVTNDIFTKEDGEFL+++ ALP
Sbjct: 71 NFSERAFTIG-IGGPVGTGKTALMLALCKVLRDKYSLAAVTNDIFTKEDGEFLIKHGALP 129
Query: 700 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKTDILLCESGGDNLAANFSRELADY 521
EERIRAVETGGCPHAAIREDISINLGPLEELSNLYK DILLCESGGDNLAANFSRELADY
Sbjct: 130 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKADILLCESGGDNLAANFSRELADY 189
Query: 520 IIYIIDVSGGDKIPRKGGPGITQADLLVINKTDLAQAIGADLGVMERDSLRMRDGGPFVF 341
IIYIIDVS GDKIPRKGGPGITQADLLVINKTDLA A+GADL VMERD+LRMRDGGPFVF
Sbjct: 190 IIYIIDVSAGDKIPRKGGPGITQADLLVINKTDLAPAVGADLSVMERDALRMRDGGPFVF 249
Query: 340 AQVKHKVGVEEIVNHVIQAWEAATGKKR 257
AQVKH VGVE+IVNH++QAWE +TG K+
Sbjct: 250 AQVKHGVGVEDIVNHILQAWEVSTGNKK 277
Score = 111 bits (278), Expect(2) = e-127
Identities = 49/76 (64%), Positives = 59/76 (77%)
Frame = -3
Query: 1088 HHHSDHGHDHDHHDNSHSHSDGVASAGSDSYVGPDGKVYHSHDGLAPHSHEPIYSPGFFT 909
HH DH H H HH++ H+H D A+ S+VG DGKVYHSHDGLAPHSHEPIYSPG+F+
Sbjct: 6 HHMHDHDHGHHHHNHDHTHEDTKAT----SWVGADGKVYHSHDGLAPHSHEPIYSPGYFS 61
Query: 908 RRAPPLINRAFQ*KSF 861
RRAPPLI+R F ++F
Sbjct: 62 RRAPPLIDRNFSERAF 77
>ref|NP_180994.1| putative urease accessory protein; protein id: At2g34470.1,
supported by cDNA: 18513. [Arabidopsis thaliana]
gi|7488416|pir||T02334 probable urease accessory protein
[imported] - Arabidopsis thaliana
gi|3128220|gb|AAC26700.1| putative urease accessory
protein [Arabidopsis thaliana]
gi|20197163|gb|AAM14950.1| putative urease accessory
protein [Arabidopsis thaliana]
gi|21553947|gb|AAM63028.1| putative urease accessory
protein [Arabidopsis thaliana]
gi|26453270|dbj|BAC43708.1| putative urease accessory
protein [Arabidopsis thaliana]
gi|28950833|gb|AAO63340.1| At2g34470 [Arabidopsis
thaliana]
Length = 275
Score = 369 bits (948), Expect(2) = e-126
Identities = 185/208 (88%), Positives = 195/208 (92%)
Frame = -1
Query: 880 HFNERAFTIGRSAGPVGTGKTALMLALCEFLRDNYSLAAVTNDIFTKEDGEFLVRNKALP 701
+F+ERAFT+G GPVGTGKTALMLALC FLRD YSLAAVTNDIFTKEDGEFLV+N ALP
Sbjct: 68 NFSERAFTVG-IGGPVGTGKTALMLALCRFLRDKYSLAAVTNDIFTKEDGEFLVKNGALP 126
Query: 700 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKTDILLCESGGDNLAANFSRELADY 521
EERIRAVETGGCPHAAIREDISINLGPLEELSNL+K D+LLCESGGDNLAANFSRELADY
Sbjct: 127 EERIRAVETGGCPHAAIREDISINLGPLEELSNLFKADLLLCESGGDNLAANFSRELADY 186
Query: 520 IIYIIDVSGGDKIPRKGGPGITQADLLVINKTDLAQAIGADLGVMERDSLRMRDGGPFVF 341
IIYIIDVS GDKIPRKGGPGITQADLLVINKTDLA A+GADL VMERDSLRMRDGGPFVF
Sbjct: 187 IIYIIDVSAGDKIPRKGGPGITQADLLVINKTDLAAAVGADLSVMERDSLRMRDGGPFVF 246
Query: 340 AQVKHKVGVEEIVNHVIQAWEAATGKKR 257
AQVKH +GVEEIVNHV+ +WE ATGKKR
Sbjct: 247 AQVKHGLGVEEIVNHVMHSWEHATGKKR 274
Score = 107 bits (267), Expect(2) = e-126
Identities = 48/78 (61%), Positives = 55/78 (69%)
Frame = -3
Query: 1094 HHHHHSDHGHDHDHHDNSHSHSDGVASAGSDSYVGPDGKVYHSHDGLAPHSHEPIYSPGF 915
HHHHH DH HDH+ D G S+VG DGKVYHSHDGLAPHSHEPIYSPG+
Sbjct: 6 HHHHHHDHEHDHEKSDGGE---------GKASWVGKDGKVYHSHDGLAPHSHEPIYSPGY 56
Query: 914 FTRRAPPLINRAFQ*KSF 861
F+RRAPPL +R F ++F
Sbjct: 57 FSRRAPPLHDRNFSERAF 74
>emb|CAC33002.1| urease accessory protein G [Solanum tuberosum]
Length = 278
Score = 364 bits (935), Expect(2) = e-126
Identities = 181/208 (87%), Positives = 195/208 (93%)
Frame = -1
Query: 880 HFNERAFTIGRSAGPVGTGKTALMLALCEFLRDNYSLAAVTNDIFTKEDGEFLVRNKALP 701
+F+ERAFTIG GPVGTGKTALMLALC+ LR+ YSLAAVTNDIFTKEDGEFL+++ ALP
Sbjct: 71 NFSERAFTIG-IGGPVGTGKTALMLALCKVLREKYSLAAVTNDIFTKEDGEFLIKHGALP 129
Query: 700 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKTDILLCESGGDNLAANFSRELADY 521
EERIRAVETGGCPHAAIREDISINLGPLEELSNLYK DILLCESGGDNLAANFSRELADY
Sbjct: 130 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKADILLCESGGDNLAANFSRELADY 189
Query: 520 IIYIIDVSGGDKIPRKGGPGITQADLLVINKTDLAQAIGADLGVMERDSLRMRDGGPFVF 341
IIYIIDVS GDKIPRKGGPGITQADLLVINKTDLA A+GADL VMERD+LRMRDGGPFVF
Sbjct: 190 IIYIIDVSAGDKIPRKGGPGITQADLLVINKTDLAAAVGADLSVMERDALRMRDGGPFVF 249
Query: 340 AQVKHKVGVEEIVNHVIQAWEAATGKKR 257
AQVKH VGVE+IVNH++QAWE +TG K+
Sbjct: 250 AQVKHGVGVEDIVNHILQAWEVSTGNKK 277
Score = 110 bits (276), Expect(2) = e-126
Identities = 48/76 (63%), Positives = 58/76 (76%)
Frame = -3
Query: 1088 HHHSDHGHDHDHHDNSHSHSDGVASAGSDSYVGPDGKVYHSHDGLAPHSHEPIYSPGFFT 909
HH DH H H HH++ H+H D A+ S++G DGKVYHSHDGLAPHSHEPIYSPG+F+
Sbjct: 6 HHMHDHDHGHHHHNHDHTHEDTKAT----SWIGADGKVYHSHDGLAPHSHEPIYSPGYFS 61
Query: 908 RRAPPLINRAFQ*KSF 861
RRAPPLI R F ++F
Sbjct: 62 RRAPPLIERNFSERAF 77
>gb|AAD44338.1|AF160197_1 Ni-binding urease accessory protein UreG [Glycine max]
Length = 274
Score = 353 bits (906), Expect(2) = e-125
Identities = 178/197 (90%), Positives = 188/197 (95%)
Frame = -1
Query: 880 HFNERAFTIGRSAGPVGTGKTALMLALCEFLRDNYSLAAVTNDIFTKEDGEFLVRNKALP 701
+FNERAFT+G GPVGTGKTALMLALCE LR+NYSLAAVTNDIFTKEDGEFLV++KALP
Sbjct: 78 NFNERAFTVG-IGGPVGTGKTALMLALCELLRENYSLAAVTNDIFTKEDGEFLVKHKALP 136
Query: 700 EERIRAVETGGCPHAAIREDISINLGPLEELSNLYKTDILLCESGGDNLAANFSRELADY 521
EERIRAVETGGCPHAAIREDISINLGPLEELSNL+K DILLCESGGDNLAANFSRELADY
Sbjct: 137 EERIRAVETGGCPHAAIREDISINLGPLEELSNLFKADILLCESGGDNLAANFSRELADY 196
Query: 520 IIYIIDVSGGDKIPRKGGPGITQADLLVINKTDLAQAIGADLGVMERDSLRMRDGGPFVF 341
IIYIIDVSGGDKIPRKGGPGITQADLLVINKTDLA AIGADL VM+RD+LRMRDGGPFVF
Sbjct: 197 IIYIIDVSGGDKIPRKGGPGITQADLLVINKTDLAPAIGADLAVMQRDALRMRDGGPFVF 256
Query: 340 AQVKHKVGVEEIVNHVI 290
AQVKHK+GVEEI N V+
Sbjct: 257 AQVKHKIGVEEIGNLVL 273
Score = 120 bits (300), Expect(2) = e-125
Identities = 53/79 (67%), Positives = 61/79 (77%), Gaps = 1/79 (1%)
Frame = -3
Query: 1094 HHHHHSDHGHDHDHHDNS-HSHSDGVASAGSDSYVGPDGKVYHSHDGLAPHSHEPIYSPG 918
HHHHH HDH HHD+ H H DG ++S+VG DGKVYHSHDGLAPHSHEPIYSPG
Sbjct: 8 HHHHHHHQDHDHHHHDHDHHHHHDG--EGETNSWVGKDGKVYHSHDGLAPHSHEPIYSPG 65
Query: 917 FFTRRAPPLINRAFQ*KSF 861
+FTRRAPPL+NR F ++F
Sbjct: 66 YFTRRAPPLLNRNFNERAF 84
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,041,351,452
Number of Sequences: 1393205
Number of extensions: 25530428
Number of successful extensions: 111647
Number of sequences better than 10.0: 673
Number of HSP's better than 10.0 without gapping: 75006
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 96215
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 65614730658
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)