Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
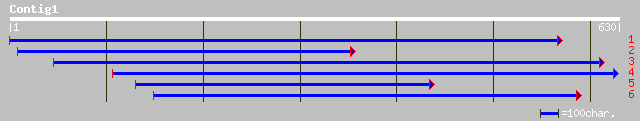
Query= KMC005070A_C01 KMC005070A_c01
(622 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181209.1| pectinesterase family; protein id: At2g36710.1 ... 32 0.002
gb|AAO22722.1| putative pectinesterase family protein [Arabidops... 32 0.002
sp|P40349|URB1_USTMA Siderophore biosynthesis regulatory protein... 33 0.004
gb|AAM44363.1| hypothetical protein [Dictyostelium discoideum] g... 31 0.005
pir||A56235 transcription activator MafB - chicken gi|516724|dbj... 33 0.006
>ref|NP_181209.1| pectinesterase family; protein id: At2g36710.1 [Arabidopsis
thaliana] gi|25408502|pir||G84783 probable
pectinesterase [imported] - Arabidopsis thaliana
gi|4415916|gb|AAD20147.1| putative pectinesterase
[Arabidopsis thaliana]
Length = 407
Score = 32.3 bits (72), Expect(2) = 0.002
Identities = 14/43 (32%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Frame = +3
Query: 201 LKTIEI-KQKDIFALAHTVFTLIQLSSSILHHHHNLHRHHYRH 326
L TI++ + + ++A T+ L + S+ H HH+ H HH+ H
Sbjct: 20 LYTIQLPSSRSLSSIATTIGALAKYSTFFGHKHHHHHHHHHHH 62
Score = 30.4 bits (67), Expect(2) = 0.002
Identities = 10/24 (41%), Positives = 12/24 (49%)
Frame = +1
Query: 400 HHN*HHHHHHQPCSLPPLNTSSHW 471
HH+ HHHHHH P+ W
Sbjct: 54 HHHHHHHHHHHYHHHEPIKCCEKW 77
>gb|AAO22722.1| putative pectinesterase family protein [Arabidopsis thaliana]
Length = 407
Score = 32.3 bits (72), Expect(2) = 0.002
Identities = 14/43 (32%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Frame = +3
Query: 201 LKTIEI-KQKDIFALAHTVFTLIQLSSSILHHHHNLHRHHYRH 326
L TI++ + + ++A T+ L + S+ H HH+ H HH+ H
Sbjct: 20 LYTIQLPSSRSLSSIATTIGALAKYSTFFGHKHHHHHHHHHHH 62
Score = 30.4 bits (67), Expect(2) = 0.002
Identities = 10/24 (41%), Positives = 12/24 (49%)
Frame = +1
Query: 400 HHN*HHHHHHQPCSLPPLNTSSHW 471
HH+ HHHHHH P+ W
Sbjct: 54 HHHHHHHHHHHYHHHEPIKCCEKW 77
>sp|P40349|URB1_USTMA Siderophore biosynthesis regulatory protein URBS1
gi|283372|pir||S27473 URBS1 protein - smut fungus
(Ustilago maydis) gi|170596|gb|AAB05617.1| urbs1
Length = 950
Score = 32.7 bits (73), Expect(2) = 0.004
Identities = 13/24 (54%), Positives = 15/24 (62%)
Frame = +1
Query: 358 ASHSPTQNQGLM*PHHN*HHHHHH 429
AS S G + PHH+ HHHHHH
Sbjct: 739 ASASDGARLGSVVPHHHHHHHHHH 762
Score = 30.4 bits (67), Expect(2) = 0.045
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +3
Query: 267 QLSSSILHHHHNLHRHHYRH 326
+L S + HHHH+ H HH H
Sbjct: 746 RLGSVVPHHHHHHHHHHANH 765
Score = 29.3 bits (64), Expect(2) = 0.004
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = +1
Query: 403 HN*HHHHHHQPCSLPPLNTSSH 468
H+ HHHHHH P + ++ SSH
Sbjct: 771 HHGHHHHHHHPVT---VSASSH 789
Score = 27.7 bits (60), Expect(2) = 0.045
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH HHHHHH
Sbjct: 771 HHGHHHHHHH 780
>gb|AAM44363.1| hypothetical protein [Dictyostelium discoideum]
gi|28828387|gb|AAM09303.2| similar to Plasmodium
lophurae. Histidine-rich glycoprotein precursor
[Dictyostelium discoideum]
Length = 233
Score = 31.2 bits (69), Expect(2) = 0.011
Identities = 11/23 (47%), Positives = 13/23 (55%)
Frame = +1
Query: 400 HHN*HHHHHHQPCSLPPLNTSSH 468
HH+ HHHHHH P P + H
Sbjct: 144 HHHHHHHHHHHPHHHPHPHPHPH 166
Score = 30.8 bits (68), Expect(2) = 0.014
Identities = 9/12 (75%), Positives = 10/12 (83%)
Frame = +1
Query: 400 HHN*HHHHHHQP 435
HH+ HHHHHH P
Sbjct: 86 HHHHHHHHHHHP 97
Score = 30.8 bits (68), Expect(2) = 0.005
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +3
Query: 285 LHHHHNLHRHHYRH 326
LHHHH+ H HH+ H
Sbjct: 70 LHHHHHHHHHHHHH 83
Score = 30.8 bits (68), Expect(2) = 0.005
Identities = 9/11 (81%), Positives = 10/11 (90%)
Frame = +1
Query: 397 PHHN*HHHHHH 429
PHH+ HHHHHH
Sbjct: 110 PHHHHHHHHHH 120
Score = 30.4 bits (67), Expect(2) = 0.018
Identities = 14/32 (43%), Positives = 15/32 (46%)
Frame = +1
Query: 346 PFTSASHSPTQNQGLM*PHHN*HHHHHHQPCS 441
P H P N PH + HHHHHHQ S
Sbjct: 185 PHPHPHHHPNPNPHPH-PHPHPHHHHHHQEAS 215
Score = 29.3 bits (64), Expect(2) = 3.7
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 135 HHHHHHHHHHHHH 147
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 116 HHHHHHHHHHHHH 128
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 73 HHHHHHHHHHHHH 85
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 115 HHHHHHHHHHHHH 127
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 76 HHHHHHHHHHHHH 88
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 112 HHHHHHHHHHHHH 124
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 82 HHHHHHHHHHHHH 94
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 80 HHHHHHHHHHHHH 92
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 72 HHHHHHHHHHHHH 84
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 118 HHHHHHHHHHHHH 130
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 84 HHHHHHHHHHHHH 96
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 121 HHHHHHHHHHHHH 133
Score = 29.3 bits (64), Expect(2) = 0.011
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 128 HHHHHHHHHHHHH 140
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 78 HHHHHHHHHHHHH 90
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 79 HHHHHHHHHHHHH 91
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 74 HHHHHHHHHHHHH 86
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 83 HHHHHHHHHHHHH 95
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 113 HHHHHHHHHHHHH 125
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 75 HHHHHHHHHHHHH 87
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 81 HHHHHHHHHHHHH 93
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 111 HHHHHHHHHHHHH 123
Score = 29.3 bits (64), Expect(2) = 0.018
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 130 HHHHHHHHHHHHH 142
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 105 HHHHHPHHHHHHH 117
Score = 29.3 bits (64), Expect(2) = 0.14
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 127 HHHHHHHHHHHHH 139
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 117 HHHHHHHHHHHHH 129
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 114 HHHHHHHHHHHHH 126
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 124 HHHHHHHHHHHHH 136
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 122 HHHHHHHHHHHHH 134
Score = 29.3 bits (64), Expect(2) = 0.014
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 77 HHHHHHHHHHHHH 89
Score = 29.3 bits (64), Expect(2) = 0.082
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 119 HHHHHHHHHHHHH 131
Score = 29.3 bits (64), Expect(2) = 3.7
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 129 HHHHHHHHHHHHH 141
Score = 28.5 bits (62), Expect(2) = 0.14
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH+ H
Sbjct: 86 HHHHHHHHHHHPH 98
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 135 HHHHHHHHHH 144
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 126 HHHHHHHHHH 135
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 117 HHHHHHHHHH 126
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 124 HHHHHHHHHH 133
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 125 HHHHHHHHHH 134
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 133 HHHHHHHHHH 142
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 121 HHHHHHHHHH 130
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 122 HHHHHHHHHH 131
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 128 HHHHHHHHHH 137
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 112 HHHHHHHHHH 121
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 123 HHHHHHHHHH 132
Score = 28.1 bits (61), Expect(2) = 0.18
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 120 HHHHHHHHHH 129
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 118 HHHHHHHHHH 127
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 83 HHHHHHHHHH 92
Score = 28.1 bits (61), Expect(2) = 1.3
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 78 HHHHHHHHHH 87
Score = 28.1 bits (61), Expect(2) = 0.18
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HH H+LH HH+ H
Sbjct: 65 HHPHHLHHHHHHH 77
Score = 28.1 bits (61), Expect(2) = 0.18
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH H HH+ H
Sbjct: 106 HHHHPHHHHHHHH 118
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 119 HHHHHHHHHH 128
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 116 HHHHHHHHHH 125
Score = 28.1 bits (61), Expect(2) = 0.49
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 130 HHHHHHHHHH 139
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 82 HHHHHHHHHH 91
Score = 28.1 bits (61), Expect(2) = 0.18
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 77 HHHHHHHHHH 86
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 134 HHHHHHHHHH 143
Score = 28.1 bits (61), Expect(2) = 0.29
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 113 HHHHHHHHHH 122
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 115 HHHHHHHHHH 124
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 131 HHHHHHHHHH 140
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 132 HHHHHHHHHH 141
Score = 28.1 bits (61), Expect(2) = 0.14
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 107 HHHPHHHHHH 116
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 81 HHHHHHHHHH 90
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 129 HHHHHHHHHH 138
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 127 HHHHHHHHHH 136
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 114 HHHHHHHHHH 123
Score = 28.1 bits (61), Expect(2) = 0.082
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH+ HHHHHH
Sbjct: 85 HHHHHHHHHH 94
Score = 27.7 bits (60), Expect(2) = 2.2
Identities = 10/14 (71%), Positives = 10/14 (71%), Gaps = 1/14 (7%)
Frame = +1
Query: 397 PHHN*HH-HHHHQP 435
PHH HH HHHH P
Sbjct: 97 PHHPHHHPHHHHHP 110
Score = 27.3 bits (59), Expect(2) = 0.14
Identities = 10/23 (43%), Positives = 12/23 (51%)
Frame = +1
Query: 400 HHN*HHHHHHQPCSLPPLNTSSH 468
HH+ HHH HH P P + H
Sbjct: 148 HHHHHHHPHHHPHPHPHPHPHPH 170
Score = 27.3 bits (59), Expect(2) = 0.29
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH H
Sbjct: 87 HHHHHHHHHHPHH 99
Score = 26.9 bits (58), Expect(2) = 3.7
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = +1
Query: 400 HHN*HHHHHH 429
HH HHHHHH
Sbjct: 108 HHPHHHHHHH 117
Score = 26.9 bits (58), Expect(2) = 1.0
Identities = 8/11 (72%), Positives = 9/11 (81%)
Frame = +1
Query: 397 PHHN*HHHHHH 429
PHH+ H HHHH
Sbjct: 104 PHHHHHPHHHH 114
Score = 26.6 bits (57), Expect(2) = 0.49
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH H HH+ H
Sbjct: 93 HHHHPHHPHHHPH 105
Score = 26.6 bits (57), Expect(2) = 1.0
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H HH H
Sbjct: 90 HHHHHHHPHHPHH 102
Score = 25.0 bits (53), Expect(2) = 1.3
Identities = 7/13 (53%), Positives = 9/13 (68%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HH H+ H HH+ H
Sbjct: 68 HHLHHHHHHHHHH 80
Score = 24.6 bits (52), Expect(2) = 3.7
Identities = 7/13 (53%), Positives = 9/13 (68%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
H HH+ H HH+ H
Sbjct: 99 HPHHHPHHHHHPH 111
Score = 24.6 bits (52), Expect(2) = 2.2
Identities = 7/13 (53%), Positives = 9/13 (68%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H H+ H
Sbjct: 89 HHHHHHHHPHHPH 101
Score = 22.3 bits (46), Expect(2) = 3.7
Identities = 9/24 (37%), Positives = 11/24 (45%)
Frame = +1
Query: 397 PHHN*HHHHHHQPCSLPPLNTSSH 468
PH + H H HH P P + H
Sbjct: 181 PHPHPHPHPHHHPNPNPHPHPHPH 204
Score = 22.3 bits (46), Expect(2) = 3.7
Identities = 10/24 (41%), Positives = 13/24 (53%)
Frame = +1
Query: 397 PHHN*HHHHHHQPCSLPPLNTSSH 468
PHH+ H H H P P L+ + H
Sbjct: 155 PHHHPHPHPHPHP--HPHLHPNPH 176
>pir||A56235 transcription activator MafB - chicken gi|516724|dbj|BAA05938.1|
MafB [Gallus gallus]
Length = 311
Score = 33.1 bits (74), Expect(2) = 0.006
Identities = 13/23 (56%), Positives = 15/23 (64%)
Frame = +1
Query: 397 PHHN*HHHHHHQPCSLPPLNTSS 465
PHH HHHHHHQ P ++SS
Sbjct: 158 PHH--HHHHHHQASPTPSTSSSS 178
Score = 29.3 bits (64), Expect(2) = 0.10
Identities = 12/27 (44%), Positives = 14/27 (51%)
Frame = +1
Query: 403 HN*HHHHHHQPCSLPPLNTSSHWKTFQ 483
H HHHHHH S P +SS + Q
Sbjct: 157 HPHHHHHHHHQASPTPSTSSSSSQQLQ 183
Score = 28.1 bits (61), Expect(2) = 0.006
Identities = 11/28 (39%), Positives = 14/28 (49%)
Frame = +3
Query: 243 AHTVFTLIQLSSSILHHHHNLHRHHYRH 326
+H V +Q S HHH+ H HH H
Sbjct: 114 SHQVSQQLQGFESFRAHHHHHHHHHQHH 141
Score = 27.7 bits (60), Expect(2) = 0.10
Identities = 7/13 (53%), Positives = 12/13 (91%)
Frame = +3
Query: 288 HHHHNLHRHHYRH 326
HHHH+ H+HH+++
Sbjct: 132 HHHHHHHQHHHQY 144
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 542,807,251
Number of Sequences: 1393205
Number of extensions: 12617685
Number of successful extensions: 109489
Number of sequences better than 10.0: 467
Number of HSP's better than 10.0 without gapping: 59715
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 92577
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)