Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005069A_C01 KMC005069A_c01
(604 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
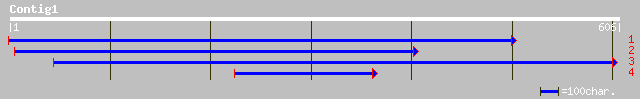
gb|AAF17704.1|AF033561_1 ornithine carbamoyltransferase [Canaval... 249 3e-65
sp|Q43814|OTC_PEA Ornithine carbamoyltransferase, chloroplast pr... 205 3e-52
gb|AAF17705.1|AF033562_1 ornithine carbamoyltransferase; OCTase ... 199 3e-50
ref|NP_177667.1| ornithine carbamoyltransferase precursor; prote... 190 1e-47
pir||T51617 ornithine carbamoyltransferase (EC 2.1.3.3) [validat... 187 7e-47
>gb|AAF17704.1|AF033561_1 ornithine carbamoyltransferase [Canavalia lineata]
gi|15428286|gb|AAK97813.1|AF203387_1 ornithine
carbamoyltransferase OOCT2 [Canavalia lineata]
Length = 369
Score = 249 bits (635), Expect = 3e-65
Identities = 134/191 (70%), Positives = 154/191 (80%), Gaps = 1/191 (0%)
Frame = +2
Query: 35 MATISHNFL-VQSDTGSFSTVILSPSYFPSSPIFRQFARFPRISTSPFPLKIACRASSVE 211
MA+IS NF +++D S S+ S S PI RQ FPR + S F L ++CRASSV+
Sbjct: 1 MASISSNFSSIRTDKASLSSAFFRSS---SVPILRQSHTFPR-NISHFTLTVSCRASSVQ 56
Query: 212 PSSVTAKTEVKDFLHINDFDKDTITKILNRAIEVKAQIKSGDRTFKPFQGMTLAMVFAKP 391
S VTAKTEVKDFLHINDFDK +I KIL+RAIEVKA IKSGDR F+PF+G T+AMVF KP
Sbjct: 57 SSPVTAKTEVKDFLHINDFDKKSIFKILDRAIEVKALIKSGDRAFQPFKGKTMAMVFCKP 116
Query: 392 SLRTHVSFETGFSWLGGHAVYLAPEDIQMGTREETRDVARIWSRFTDIIMARVFTHEDIL 571
SLRTHVSFETGF+ LGGHAVYLAP DIQMGTREETRDVARI SRFTDIIMARVF+H+DIL
Sbjct: 117 SLRTHVSFETGFTLLGGHAVYLAPTDIQMGTREETRDVARILSRFTDIIMARVFSHQDIL 176
Query: 572 TLSKHATVPVI 604
LSK+ +VPV+
Sbjct: 177 VLSKYGSVPVV 187
>sp|Q43814|OTC_PEA Ornithine carbamoyltransferase, chloroplast precursor (OTCase)
(Ornithine transcarbamylase) gi|7433603|pir||T06436
probable ornithine carbamoyltransferase (EC 2.1.3.3) -
garden pea gi|971168|gb|AAA74997.1| ornithine
carbamoyltransferase
Length = 375
Score = 205 bits (522), Expect = 3e-52
Identities = 112/185 (60%), Positives = 140/185 (75%), Gaps = 11/185 (5%)
Frame = +2
Query: 83 FSTVILSPSYFPSSPIFRQFARFPRIS------TSPFPLK-IACRASSVEP--SSVTAKT 235
F+TV Y SP F P +S +SP PL+ I+C+ASS S++TAK
Sbjct: 11 FTTVGSHKPYL--SPSSHNFRHSPSVSLSSSSSSSPSPLRRISCQASSAPAAESTLTAKV 68
Query: 236 E--VKDFLHINDFDKDTITKILNRAIEVKAQIKSGDRTFKPFQGMTLAMVFAKPSLRTHV 409
+KDF+HI+DFDK+TI KIL+RAIEVK +KSGDRTF+PF+G T++M+FAKPS+RT V
Sbjct: 69 GNGLKDFIHIDDFDKETILKILDRAIEVKTLLKSGDRTFRPFEGKTMSMIFAKPSMRTRV 128
Query: 410 SFETGFSWLGGHAVYLAPEDIQMGTREETRDVARIWSRFTDIIMARVFTHEDILTLSKHA 589
SFETGFS LGGHA+YL P DIQMG REETRDVAR+ SR+ DIIMARVF+H+DIL L+K+A
Sbjct: 129 SFETGFSLLGGHAIYLGPNDIQMGKREETRDVARVLSRYNDIIMARVFSHQDILDLAKYA 188
Query: 590 TVPVI 604
+VPVI
Sbjct: 189 SVPVI 193
>gb|AAF17705.1|AF033562_1 ornithine carbamoyltransferase; OCTase [Canavalia lineata]
gi|12003188|gb|AAG43481.1|AF203688_1 ornithine
carbamoyltransferase OOCT1 [Canavalia lineata]
Length = 359
Score = 199 bits (505), Expect = 3e-50
Identities = 102/174 (58%), Positives = 131/174 (74%)
Frame = +2
Query: 83 FSTVILSPSYFPSSPIFRQFARFPRISTSPFPLKIACRASSVEPSSVTAKTEVKDFLHIN 262
+ TV S + S R+ P +S S F +I+C A+S P V K +FLH++
Sbjct: 9 YCTVRGSDKPYLSGQNLRRRTPLPAVSVS-FRRRISCIATSAAPQKVGQK----NFLHLD 63
Query: 263 DFDKDTITKILNRAIEVKAQIKSGDRTFKPFQGMTLAMVFAKPSLRTHVSFETGFSWLGG 442
DFDKDTI K+L+RA+EVK+ +KSGDRTF+PF+G T++M+F KPS+RT VSFETGF+ LGG
Sbjct: 64 DFDKDTILKMLDRALEVKSLLKSGDRTFRPFEGKTMSMIFTKPSMRTRVSFETGFTLLGG 123
Query: 443 HAVYLAPEDIQMGTREETRDVARIWSRFTDIIMARVFTHEDILTLSKHATVPVI 604
HA+YL P+DIQMG REETRDVAR+ SR+ DIIMARVF H+DIL L+K+ATVPVI
Sbjct: 124 HAIYLGPDDIQMGKREETRDVARVLSRYNDIIMARVFAHQDILDLAKYATVPVI 177
>ref|NP_177667.1| ornithine carbamoyltransferase precursor; protein id: At1g75330.1,
supported by cDNA: gi_13878014, supported by cDNA:
gi_17104630, supported by cDNA: gi_2764517 [Arabidopsis
thaliana] gi|14917022|sp|O50039|OTC_ARATH Ornithine
carbamoyltransferase, chloroplast precursor (OTCase)
(Ornithine transcarbamylase) gi|25286265|pir||G96783
ornithine carbamoyltransferase [imported] - Arabidopsis
thaliana gi|10120450|gb|AAG13075.1|AC023754_13 Ornithine
carbamoyltransferase [Arabidopsis thaliana]
gi|13878015|gb|AAK44085.1|AF370270_1 putative ornithine
carbamoyltransferase precursor [Arabidopsis thaliana]
gi|17104631|gb|AAL34204.1| putative ornithine
carbamoyltransferase precursor [Arabidopsis thaliana]
Length = 375
Score = 190 bits (483), Expect = 1e-47
Identities = 103/193 (53%), Positives = 139/193 (71%), Gaps = 4/193 (2%)
Frame = +2
Query: 38 ATISHNFLVQSDTGSFSTVILSPSYFPSSPIFRQFARFPRISTSPFPLKIACRASSV--- 208
A SH +S SFS+ S S+FP + + R+F+ S + L+++C+ASSV
Sbjct: 4 AMASHVSTARSPALSFSSS--SSSFFPGTTL-RRFSAVSLPSPALPRLRVSCQASSVTSP 60
Query: 209 -EPSSVTAKTEVKDFLHINDFDKDTITKILNRAIEVKAQIKSGDRTFKPFQGMTLAMVFA 385
PS V K+++KDFL I+DFD TI IL++A EVKA +KSG+R + PF+G +++M+FA
Sbjct: 61 SSPSDVKGKSDLKDFLAIDDFDTATIKTILDKASEVKALLKSGERNYLPFKGKSMSMIFA 120
Query: 386 KPSLRTHVSFETGFSWLGGHAVYLAPEDIQMGTREETRDVARIWSRFTDIIMARVFTHED 565
KPS+RT VSFETGF LGGHA+YL P DIQMG REETRDVAR+ SR+ DIIMARVF H+D
Sbjct: 121 KPSMRTRVSFETGFFLLGGHALYLGPNDIQMGKREETRDVARVLSRYNDIIMARVFAHQD 180
Query: 566 ILTLSKHATVPVI 604
IL L+ +++VPV+
Sbjct: 181 ILDLANYSSVPVV 193
>pir||T51617 ornithine carbamoyltransferase (EC 2.1.3.3) [validated] -
Arabidopsis thaliana gi|2764518|emb|CAA04115.1|
Ornithine carbamoyltransferase [Arabidopsis thaliana]
gi|2764737|emb|CAA05510.1| ornithine
carbamoyltransferase [Arabidopsis thaliana]
Length = 375
Score = 187 bits (476), Expect = 7e-47
Identities = 102/193 (52%), Positives = 138/193 (70%), Gaps = 4/193 (2%)
Frame = +2
Query: 38 ATISHNFLVQSDTGSFSTVILSPSYFPSSPIFRQFARFPRISTSPFPLKIACRASSV--- 208
A SH +S SFS+ S S+FP + + R+F+ S + L+++C+ASSV
Sbjct: 4 AMASHVSTARSPALSFSSS--SSSFFPGTTL-RRFSAVSLPSPALPRLRVSCQASSVTSP 60
Query: 209 -EPSSVTAKTEVKDFLHINDFDKDTITKILNRAIEVKAQIKSGDRTFKPFQGMTLAMVFA 385
PS V K+++KDFL I+DFD TI IL++A EVKA +KSG+R + PF+G +++M+FA
Sbjct: 61 SSPSDVKGKSDLKDFLAIDDFDTATIKTILDKASEVKALLKSGERNYLPFKGKSMSMIFA 120
Query: 386 KPSLRTHVSFETGFSWLGGHAVYLAPEDIQMGTREETRDVARIWSRFTDIIMARVFTHED 565
KPS+RT VSFETGF LGGHA+YL P DIQMG REET DVAR+ SR+ DIIMARVF H+D
Sbjct: 121 KPSMRTRVSFETGFFLLGGHALYLGPNDIQMGKREETPDVARVLSRYNDIIMARVFAHQD 180
Query: 566 ILTLSKHATVPVI 604
IL L+ +++VPV+
Sbjct: 181 ILDLANYSSVPVV 193
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 536,790,327
Number of Sequences: 1393205
Number of extensions: 11732278
Number of successful extensions: 33533
Number of sequences better than 10.0: 380
Number of HSP's better than 10.0 without gapping: 32203
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33449
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23711793746
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)