Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005061A_C01 KMC005061A_c01
(1281 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
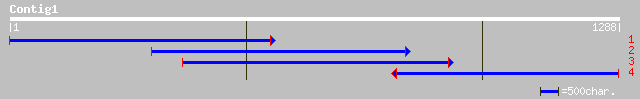
dbj|BAC22500.1| resistant specific protein-2 [Vigna radiata] 214 3e-61
dbj|BAC22501.1| resistant specific protein-3 [Vigna radiata] 211 4e-61
dbj|BAC22498.1| resistant specific protein-1(4) [Vigna radiata] ... 211 1e-60
gb|AAL67991.1| dehydration-induced protein RD22-like protein [Go... 189 6e-57
gb|AAL26909.1|AF319165_1 dehydration-responsive protein RD22 [Pr... 186 8e-57
>dbj|BAC22500.1| resistant specific protein-2 [Vigna radiata]
Length = 440
Score = 214 bits (546), Expect(2) = 3e-61
Identities = 98/170 (57%), Positives = 129/170 (75%)
Frame = +3
Query: 579 KVLEILVVKPDSKKTEIVEKTMGGCELPSMEGEEQMCATSLESLVDFIASKLGKNVLAIS 758
++LEIL VKPDSK + VEKT+ CE P+++GEE+ CATS+ES+VDF+ SKLG N S
Sbjct: 273 EILEILAVKPDSKNAKNVEKTLNNCEEPALKGEEKHCATSVESMVDFVTSKLGNNARVTS 332
Query: 759 TEVELEKETQSQNLLVKDGVKKLADDGMGVCHPMTYPYAVCFSHKLVKTSAYFVPLEGED 938
TE+E+E ++ Q +VKDGVK LA++ + CHPM+YPY V + HK+ ++A+ VPLEGED
Sbjct: 333 TELEIE--SKFQKFIVKDGVKILAEEEIIACHPMSYPYVVFYCHKMSNSTAHVVPLEGED 390
Query: 939 GARVKAVAVCHRDTSKWDPNHVSFPDLNVKPGTVPVCHVLPERHLLWVPK 1088
G RVKA+ +CH+DTS+WDP+HV+F L VKPGT PVCH P HLLW K
Sbjct: 391 GTRVKAIVICHKDTSQWDPDHVAFQVLKVKPGTSPVCHFFPNGHLLWYAK 440
Score = 44.7 bits (104), Expect(2) = 3e-61
Identities = 22/48 (45%), Positives = 29/48 (59%)
Frame = +2
Query: 437 KAGVFFFEENLHPGTKLDLQLSKRKPAVPILPRTIADQIPFSLEKLKE 580
K +F EE L G KL + KRK + P+L R IA+ +PFS EK+ E
Sbjct: 226 KPSSYFSEEGLRRGAKLVMLFHKRKFSTPLLTREIAEHLPFSSEKINE 273
Score = 39.3 bits (90), Expect = 0.14
Identities = 16/35 (45%), Positives = 25/35 (70%)
Frame = +2
Query: 44 FSLSMMVMLAYAALPPQLDWKMMLPTTPVPKAITE 148
FSL +++M A A+LP ++ W+ LP TP+PK I +
Sbjct: 10 FSLIVILMAAQASLPSEVYWERKLPNTPIPKVIRQ 44
>dbj|BAC22501.1| resistant specific protein-3 [Vigna radiata]
Length = 275
Score = 211 bits (536), Expect(2) = 4e-61
Identities = 96/170 (56%), Positives = 129/170 (75%)
Frame = +3
Query: 579 KVLEILVVKPDSKKTEIVEKTMGGCELPSMEGEEQMCATSLESLVDFIASKLGKNVLAIS 758
++LEIL VKPDSK + VE+T+ CE P+++GEE+ CATS+ES+VDF+ SKLG N S
Sbjct: 108 EILEILAVKPDSKDAKNVEETLNHCEKPALKGEEKQCATSVESMVDFVTSKLGNNARVTS 167
Query: 759 TEVELEKETQSQNLLVKDGVKKLADDGMGVCHPMTYPYAVCFSHKLVKTSAYFVPLEGED 938
TE+E+ ++ Q +VKDGVK LA++ + CHPM+YPY V + HK+ ++A+F+PLEGED
Sbjct: 168 TELEIG--SKFQKFIVKDGVKILAEEKIIACHPMSYPYVVFYCHKMANSTAHFLPLEGED 225
Query: 939 GARVKAVAVCHRDTSKWDPNHVSFPDLNVKPGTVPVCHVLPERHLLWVPK 1088
G RVKAVA+CH+DTS+WDP+HV+F L VKPGT CH PE HL+W K
Sbjct: 226 GTRVKAVAICHKDTSQWDPHHVAFQVLKVKPGTSSACHFFPEGHLVWYAK 275
Score = 50.8 bits (120), Expect = 5e-05
Identities = 23/44 (52%), Positives = 32/44 (72%)
Frame = +2
Query: 44 FSLSMMVMLAYAALPPQLDWKMMLPTTPVPKAITEQLSLGDIVS 175
FSL +++M A AALPP++ W+ MLP TP+PK I + LG IV+
Sbjct: 10 FSLIVILMAAQAALPPEVYWERMLPNTPIPKVIRQFSELGPIVN 53
Score = 47.8 bits (112), Expect(2) = 4e-61
Identities = 25/69 (36%), Positives = 36/69 (51%)
Frame = +2
Query: 374 KSVMEYKKLVLVFTSRCTQKTKAGVFFFEENLHPGTKLDLQLSKRKPAVPILPRTIADQI 553
K + ++ +L + K FF EE L G KLD+ KR + P+L R IA+ +
Sbjct: 40 KVIRQFSELGPIVNHHHHDHLKPSNFFSEERLRRGAKLDVLFRKRNFSTPLLTREIAEHL 99
Query: 554 PFSLEKLKE 580
PFS EK+ E
Sbjct: 100 PFSSEKINE 108
>dbj|BAC22498.1| resistant specific protein-1(4) [Vigna radiata]
gi|24416616|dbj|BAC22499.1| resistant specific
protein-1(8) [Vigna radiata]
Length = 402
Score = 211 bits (536), Expect(2) = 1e-60
Identities = 96/170 (56%), Positives = 129/170 (75%)
Frame = +3
Query: 579 KVLEILVVKPDSKKTEIVEKTMGGCELPSMEGEEQMCATSLESLVDFIASKLGKNVLAIS 758
++LEIL VKPDSK + VE+T+ CE P+++GEE+ CATS+ES+VDF+ SKLG N S
Sbjct: 235 EILEILAVKPDSKDAKNVEETLNHCEKPALKGEEKQCATSVESMVDFVTSKLGNNARVTS 294
Query: 759 TEVELEKETQSQNLLVKDGVKKLADDGMGVCHPMTYPYAVCFSHKLVKTSAYFVPLEGED 938
TE+E+ ++ Q +VKDGVK LA++ + CHPM+YPY V + HK+ ++A+F+PLEGED
Sbjct: 295 TELEIG--SKFQKFIVKDGVKILAEEKIIACHPMSYPYVVFYCHKMANSTAHFLPLEGED 352
Query: 939 GARVKAVAVCHRDTSKWDPNHVSFPDLNVKPGTVPVCHVLPERHLLWVPK 1088
G RVKAVA+CH+DTS+WDP+HV+F L VKPGT CH PE HL+W K
Sbjct: 353 GTRVKAVAICHKDTSQWDPHHVAFQVLKVKPGTSSACHFFPEGHLVWYAK 402
Score = 46.6 bits (109), Expect(2) = 1e-60
Identities = 23/48 (47%), Positives = 29/48 (59%)
Frame = +2
Query: 437 KAGVFFFEENLHPGTKLDLQLSKRKPAVPILPRTIADQIPFSLEKLKE 580
K FF EE L G KLD+ KR + P+L R IA+ +PFS EK+ E
Sbjct: 188 KPSNFFSEERLRRGAKLDVLFRKRNFSTPLLTREIAEHLPFSSEKINE 235
Score = 45.8 bits (107), Expect = 0.001
Identities = 19/35 (54%), Positives = 27/35 (76%)
Frame = +2
Query: 44 FSLSMMVMLAYAALPPQLDWKMMLPTTPVPKAITE 148
FSL +++M A AALPP++ W+ MLP TP+PK I +
Sbjct: 10 FSLIVILMAAQAALPPEVYWERMLPNTPIPKVIRQ 44
>gb|AAL67991.1| dehydration-induced protein RD22-like protein [Gossypium hirsutum]
Length = 335
Score = 189 bits (479), Expect(2) = 6e-57
Identities = 90/163 (55%), Positives = 114/163 (69%)
Frame = +3
Query: 600 VKPDSKKTEIVEKTMGGCELPSMEGEEQMCATSLESLVDFIASKLGKNVLAISTEVELEK 779
VKP S K E+++ T+ CE P++EGEE+ CATSLES++D+ SKLGK A+STEVE K
Sbjct: 175 VKPGSLKAEMMKNTIKECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQAVSTEVE--K 232
Query: 780 ETQSQNLLVKDGVKKLADDGMGVCHPMTYPYAVCFSHKLVKTSAYFVPLEGEDGARVKAV 959
+T Q + GV+K+ DD VCH Y YAV + HK T AY VPLEG DG + KAV
Sbjct: 233 QTPMQKYTIAAGVQKMTDDKAVVCHKQNYAYAVFYCHKSETTRAYMVPLEGADGTKAKAV 292
Query: 960 AVCHRDTSKWDPNHVSFPDLNVKPGTVPVCHVLPERHLLWVPK 1088
AVCH DTS W+P H++F L V+PGT+PVCH LP H++WVPK
Sbjct: 293 AVCHTDTSAWNPKHLAFQVLKVEPGTIPVCHFLPRDHIVWVPK 335
Score = 55.8 bits (133), Expect(2) = 6e-57
Identities = 49/181 (27%), Positives = 77/181 (42%), Gaps = 1/181 (0%)
Frame = +2
Query: 41 LFSLSMMVMLAYAALPPQLDWKMMLPTTPVPKAITEQLSLGDIVSRGKSVN-DAETQSQN 217
L L++ V++++AAL P+ W LP TP+PKA+ E L + + SVN + N
Sbjct: 8 LACLALAVVVSHAALSPEQYWSYKLPNTPMPKAVKEILHPELMEEKSTSVNVGGGGVNVN 67
Query: 218 MFHGF*GRFHHNEIGSCFGCNVHSSFLVYRHNILYGV*TLCAKADPSFMHDTKSVMEYKK 397
G G H +G G V++ G T DP +
Sbjct: 68 TGKGKPGGDTHVNVGG-KGVGVNTG--------KPGGGTHVNVGDP-----------FNY 107
Query: 398 LVLVFTSRCTQKTKAGVFFFEENLHPGTKLDLQLSKRKPAVPILPRTIADQIPFSLEKLK 577
L ++ + +FF E+++HPG + L ++ LP A +IPFS +KL
Sbjct: 108 LYAASETQIHEDPNVALFFLEKDMHPGATMSLHFTENTEKSAFLPYQTAQKIPFSSDKLP 167
Query: 578 E 580
E
Sbjct: 168 E 168
>gb|AAL26909.1|AF319165_1 dehydration-responsive protein RD22 [Prunus persica]
Length = 349
Score = 186 bits (473), Expect(2) = 8e-57
Identities = 93/166 (56%), Positives = 113/166 (68%)
Frame = +3
Query: 600 VKPDSKKTEIVEKTMGGCELPSMEGEEQMCATSLESLVDFIASKLGKNVLAISTEVELEK 779
VKP+S + +I++ T+ CE + GEE+ CATSLES+VDF SKLG N+ AISTE E K
Sbjct: 186 VKPESVEADIIKGTIEECESSGIRGEEKYCATSLESMVDFSTSKLGGNIQAISTEAE--K 243
Query: 780 ETQSQNLLVKDGVKKLADDGMGVCHPMTYPYAVCFSHKLVKTSAYFVPLEGEDGARVKAV 959
Q + GVKKLA VCH TYPYAV + H T AY VPL+G DG KAV
Sbjct: 244 GATLQKYTITPGVKKLAAGKSVVCHKQTYPYAVFYCHATKTTRAYVVPLKGADGLEAKAV 303
Query: 960 AVCHRDTSKWDPNHVSFPDLNVKPGTVPVCHVLPERHLLWVPK*ST 1097
AVCH DTS+W+P H++F L VKPGTVPVCH LP+ H++WVPK ST
Sbjct: 304 AVCHTDTSEWNPKHLAFQVLKVKPGTVPVCHFLPKDHIVWVPKKST 349
Score = 57.8 bits (138), Expect(2) = 8e-57
Identities = 49/181 (27%), Positives = 76/181 (41%), Gaps = 8/181 (4%)
Frame = +2
Query: 62 VMLAYAALPPQLDWKMMLPTTPVPKAITEQLSLGDIVSRGKSVN----DAETQSQNMFHG 229
++ ++AALPPQL W +LP T +PK+I+E L +G +V Q+ G
Sbjct: 1 LVASHAALPPQLYWNSVLPNTQMPKSISELLQPDFAEEKGTTVGVGKGGVNVQAGKGKPG 60
Query: 230 F*GRFHHNEIGSCFGCNVHSSFLVYRHNILYGV*TLCA----KADPSFMHDTKSVMEYKK 397
G + N G G V + N+ G + K P ++ +
Sbjct: 61 --GGTNVNVGGKGGGVGVTTGKPGGGTNVGVGKGGVSVNTGHKGKPVYVGVKPGPNPFNY 118
Query: 398 LVLVFTSRCTQKTKAGVFFFEENLHPGTKLDLQLSKRKPAVPILPRTIADQIPFSLEKLK 577
+ ++ A +FF E+++ PGT + L S +PR AD IPFS KL
Sbjct: 119 VYAATETQLHGNRNAAIFFLEKDIRPGTSMTLTFSGNSNTAAFVPRKTADSIPFSSNKLP 178
Query: 578 E 580
E
Sbjct: 179 E 179
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,137,066,385
Number of Sequences: 1393205
Number of extensions: 25675345
Number of successful extensions: 58782
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 55492
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 58718
length of database: 448,689,247
effective HSP length: 126
effective length of database: 273,145,417
effective search space used: 81943625100
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)