Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005047A_C01 KMC005047A_c01
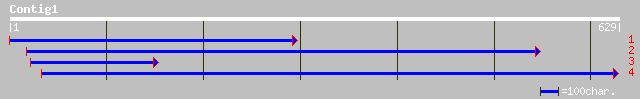
(621 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO13749.1|AF268257_1 PTEN-like protein [Arabidopsis thaliana] 90 2e-17
ref|NP_188573.1| putative tyrosine phosphatase; protein id: At3g... 90 2e-17
ref|NP_566929.1| putative tyrosine phosphatase; protein id: At3g... 58 1e-07
pir||T45864 probable tyrosine phosphatase - Arabidopsis thaliana... 58 1e-07
pir||T46707 proteophosphoglycan, membrane-associated [imported] ... 45 6e-04
>gb|AAO13749.1|AF268257_1 PTEN-like protein [Arabidopsis thaliana]
Length = 611
Score = 90.1 bits (222), Expect = 2e-17
Identities = 56/140 (40%), Positives = 85/140 (60%), Gaps = 7/140 (5%)
Frame = -3
Query: 604 APVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRSKQTKASEAVETVTNAPRESESN 425
+PV+ S + P KE+E +KDDVFSD E + +++ + +S+ E+
Sbjct: 484 SPVDGSASVPGPDKETENPDKDDVFSDNEGDSTGPTKTTSSASSQT----------PEAK 533
Query: 424 KNSDQVSNLTRATEQVSL-GNKIST-PIHSAGEPKSD-----DGRTVPSLPTPSSESEFK 266
K++D+ + LT+ATE+VS+ GNK S+ P+ K + G V + + SSESEFK
Sbjct: 534 KSADETAVLTKATEKVSISGNKGSSQPVQGVTVSKGEATEKPSGAGVNA--SSSSESEFK 591
Query: 265 AMAADASVFTFGDDEDYESD 206
MAADASVF+FGD++D+ESD
Sbjct: 592 VMAADASVFSFGDEDDFESD 611
>ref|NP_188573.1| putative tyrosine phosphatase; protein id: At3g19420.1, supported
by cDNA: gi_17979062 [Arabidopsis thaliana]
gi|11994464|dbj|BAB02466.1| tyrosine phosphatase-like
protein [Arabidopsis thaliana]
gi|17979063|gb|AAL49799.1| putative tyrosine phosphatase
[Arabidopsis thaliana]
Length = 611
Score = 90.1 bits (222), Expect = 2e-17
Identities = 56/140 (40%), Positives = 85/140 (60%), Gaps = 7/140 (5%)
Frame = -3
Query: 604 APVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRSKQTKASEAVETVTNAPRESESN 425
+PV+ S + P KE+E +KDDVFSD E + +++ + +S+ E+
Sbjct: 484 SPVDGSASVPGPDKETENPDKDDVFSDNEGDSTGPTKTTSSASSQT----------PEAK 533
Query: 424 KNSDQVSNLTRATEQVSL-GNKIST-PIHSAGEPKSD-----DGRTVPSLPTPSSESEFK 266
K++D+ + LT+ATE+VS+ GNK S+ P+ K + G V + + SSESEFK
Sbjct: 534 KSADETAVLTKATEKVSISGNKGSSQPVQGVTVSKGEATEKPSGAGVNA--SSSSESEFK 591
Query: 265 AMAADASVFTFGDDEDYESD 206
MAADASVF+FGD++D+ESD
Sbjct: 592 VMAADASVFSFGDEDDFESD 611
>ref|NP_566929.1| putative tyrosine phosphatase; protein id: At3g50110.1, supported
by cDNA: gi_16604644 [Arabidopsis thaliana]
gi|24030328|gb|AAN41331.1| putative tyrosine phosphatase
[Arabidopsis thaliana]
Length = 632
Score = 57.8 bits (138), Expect = 1e-07
Identities = 45/140 (32%), Positives = 69/140 (49%), Gaps = 3/140 (2%)
Frame = -3
Query: 616 SNSPAPVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRSKQT--KASEAVETVTNAP 443
S+S S ++ ++SK + DDVFSD + E +S+S T K + ++ T +
Sbjct: 512 SDSTQQQSQSSSSADSSKLKSNEKDDDVFSDSDGEEEGNSQSYSTNEKTASSMHTTSKPH 571
Query: 442 RESESNKNSDQVSNLTRATEQVSLGNKISTPIHSAGEPKSDDGRTVPSLPTPSSESEFKA 263
+ +E K D +N + T S G+ P +S + S+ KA
Sbjct: 572 QINEPPKRDDPSANRS-VTSSSSSGHYNPIPNNSL------------------AVSDIKA 612
Query: 262 MAADASVFTFGD-DEDYESD 206
+AADASVF+FGD +EDYESD
Sbjct: 613 IAADASVFSFGDEEEDYESD 632
>pir||T45864 probable tyrosine phosphatase - Arabidopsis thaliana
gi|6522932|emb|CAB62119.1| putative tyrosine phosphatase
[Arabidopsis thaliana]
Length = 628
Score = 57.8 bits (138), Expect = 1e-07
Identities = 45/140 (32%), Positives = 69/140 (49%), Gaps = 3/140 (2%)
Frame = -3
Query: 616 SNSPAPVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRSKQT--KASEAVETVTNAP 443
S+S S ++ ++SK + DDVFSD + E +S+S T K + ++ T +
Sbjct: 508 SDSTQQQSQSSSSADSSKLKSNEKDDDVFSDSDGEEEGNSQSYSTNEKTASSMHTTSKPH 567
Query: 442 RESESNKNSDQVSNLTRATEQVSLGNKISTPIHSAGEPKSDDGRTVPSLPTPSSESEFKA 263
+ +E K D +N + T S G+ P +S + S+ KA
Sbjct: 568 QINEPPKRDDPSANRS-VTSSSSSGHYNPIPNNSL------------------AVSDIKA 608
Query: 262 MAADASVFTFGD-DEDYESD 206
+AADASVF+FGD +EDYESD
Sbjct: 609 IAADASVFSFGDEEEDYESD 628
>pir||T46707 proteophosphoglycan, membrane-associated [imported] - Leishmania
major (fragment) gi|5420389|emb|CAB46680.1|
proteophosphoglycan [Leishmania major]
Length = 383
Score = 45.4 bits (106), Expect = 6e-04
Identities = 34/125 (27%), Positives = 54/125 (43%), Gaps = 1/125 (0%)
Frame = -3
Query: 616 SNSPAPVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRS-KQTKASEAVETVTNAPR 440
S+S AP S +AP+AS S + S + P+SS S +S A + ++AP
Sbjct: 60 SSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPS 119
Query: 439 ESESNKNSDQVSNLTRATEQVSLGNKISTPIHSAGEPKSDDGRTVPSLPTPSSESEFKAM 260
S S+ S S + ++ + S P S+ S + PS + S+ S +
Sbjct: 120 ASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSS 179
Query: 259 AADAS 245
A AS
Sbjct: 180 APSAS 184
Score = 40.8 bits (94), Expect = 0.014
Identities = 33/140 (23%), Positives = 57/140 (40%), Gaps = 10/140 (7%)
Frame = -3
Query: 616 SNSPAPVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRSKQTKASEAVETV------ 455
S+S AP S +AP++S + A S + ASS S + +S + +
Sbjct: 98 SSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAP 157
Query: 454 ----TNAPRESESNKNSDQVSNLTRATEQVSLGNKISTPIHSAGEPKSDDGRTVPSLPTP 287
++AP S S+ S S+ A+ + + S P S+ S + PS +
Sbjct: 158 SSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSS 217
Query: 286 SSESEFKAMAADASVFTFGD 227
S+ S + + +S T D
Sbjct: 218 SAPSSSSSAPSSSSTTTTMD 237
Score = 39.7 bits (91), Expect = 0.032
Identities = 30/126 (23%), Positives = 53/126 (41%), Gaps = 2/126 (1%)
Frame = -3
Query: 616 SNSPAPVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRSKQTKASEAVETV--TNAP 443
S+S AP S +AP++S + A S + ASS S + +S + + ++AP
Sbjct: 6 SSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAP 65
Query: 442 RESESNKNSDQVSNLTRATEQVSLGNKISTPIHSAGEPKSDDGRTVPSLPTPSSESEFKA 263
S S+ S S+ ++ + S P S+ P + S + S S A
Sbjct: 66 SSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSA 125
Query: 262 MAADAS 245
++ +S
Sbjct: 126 PSSSSS 131
Score = 36.2 bits (82), Expect = 0.35
Identities = 34/125 (27%), Positives = 53/125 (42%), Gaps = 1/125 (0%)
Frame = -3
Query: 616 SNSPAPVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRS-KQTKASEAVETVTNAPR 440
S+S AP S +AP+AS S + S + P+SS S +S A + ++AP
Sbjct: 76 SSSSAPSS-SSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPS 134
Query: 439 ESESNKNSDQVSNLTRATEQVSLGNKISTPIHSAGEPKSDDGRTVPSLPTPSSESEFKAM 260
S S+ S S+ A+ S P S+ S + PS + S+ S +
Sbjct: 135 ASSSSAPSSSSSSAPSASSS-------SAPSSSSSSAPSASSSSAPSSSSSSAPSASSSS 187
Query: 259 AADAS 245
A +S
Sbjct: 188 APSSS 192
Score = 36.2 bits (82), Expect = 0.35
Identities = 31/133 (23%), Positives = 54/133 (40%), Gaps = 9/133 (6%)
Frame = -3
Query: 616 SNSPAPVEVSPAAPNASKESEGAEKDDVFSDGEAEHPASSRSKQTKASEA---------V 464
S+S AP S +AP++S + A S + ASS S + +S A
Sbjct: 83 SSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPS 142
Query: 463 ETVTNAPRESESNKNSDQVSNLTRATEQVSLGNKISTPIHSAGEPKSDDGRTVPSLPTPS 284
+ ++AP S S+ S S+ A+ + + S+ ++ + PS + S
Sbjct: 143 SSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSS 202
Query: 283 SESEFKAMAADAS 245
+ S + A AS
Sbjct: 203 APSSSSSSAPSAS 215
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 509,236,962
Number of Sequences: 1393205
Number of extensions: 10382719
Number of successful extensions: 42611
Number of sequences better than 10.0: 287
Number of HSP's better than 10.0 without gapping: 39170
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42275
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)