Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005038A_C01 KMC005038A_c01
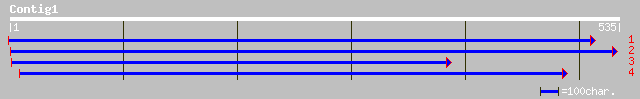
(531 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T49014 hypothetical protein F3C22.10 - Arabidopsis thaliana... 174 8e-43
gb|AAK59422.1| unknown protein [Arabidopsis thaliana] 174 8e-43
ref|NP_566970.1| expressed protein; protein id: At3g52610.1, sup... 174 8e-43
dbj|BAB63795.1| P0423B08.4 [Oryza sativa (japonica cultivar-grou... 143 1e-33
ref|NP_494910.1| Trypsin precursor [Caenorhabditis elegans] gi|7... 35 0.72
>pir||T49014 hypothetical protein F3C22.10 - Arabidopsis thaliana
gi|7669935|emb|CAB89222.1| putative protein [Arabidopsis
thaliana]
Length = 465
Score = 174 bits (440), Expect = 8e-43
Identities = 83/117 (70%), Positives = 98/117 (82%), Gaps = 1/117 (0%)
Frame = -2
Query: 524 NSNKHFSDMNDLNFLMCKAIENPGLTPSSSLRTALISVFEDAVIDDSSQVH-EELGLEDY 348
NSNK F+DM+DLNFLMCKAIENP LTPSSSLRTA IS+FED VID+S + LG++DY
Sbjct: 347 NSNKQFTDMSDLNFLMCKAIENPNLTPSSSLRTAFISIFEDPVIDESPEPELASLGVQDY 406
Query: 347 VGCASTHGVGPSVAIFDTVRNGKLDCACIYPSPLHSREQIQGLVDDMKRILVEGCNS 177
+GCAS HGVGPSVA+FD +R+GKLDCA +YPSPLHSREQ+ GL+ MK IL+EG S
Sbjct: 407 IGCASIHGVGPSVAVFDALRDGKLDCAFVYPSPLHSREQMDGLIQHMKTILLEGSAS 463
>gb|AAK59422.1| unknown protein [Arabidopsis thaliana]
Length = 475
Score = 174 bits (440), Expect = 8e-43
Identities = 83/117 (70%), Positives = 98/117 (82%), Gaps = 1/117 (0%)
Frame = -2
Query: 524 NSNKHFSDMNDLNFLMCKAIENPGLTPSSSLRTALISVFEDAVIDDSSQVH-EELGLEDY 348
NSNK F+DM+DLNFLMCKAIENP LTPSSSLRTA IS+FED VID+S + LG++DY
Sbjct: 357 NSNKQFTDMSDLNFLMCKAIENPNLTPSSSLRTAFISIFEDPVIDESPEPELASLGVQDY 416
Query: 347 VGCASTHGVGPSVAIFDTVRNGKLDCACIYPSPLHSREQIQGLVDDMKRILVEGCNS 177
+GCAS HGVGPSVA+FD +R+GKLDCA +YPSPLHSREQ+ GL+ MK IL+EG S
Sbjct: 417 IGCASIHGVGPSVAVFDALRDGKLDCAFVYPSPLHSREQMDGLIQHMKTILLEGSAS 473
>ref|NP_566970.1| expressed protein; protein id: At3g52610.1, supported by cDNA:
gi_14334447 [Arabidopsis thaliana]
gi|23296330|gb|AAN13043.1| unknown protein [Arabidopsis
thaliana]
Length = 475
Score = 174 bits (440), Expect = 8e-43
Identities = 83/117 (70%), Positives = 98/117 (82%), Gaps = 1/117 (0%)
Frame = -2
Query: 524 NSNKHFSDMNDLNFLMCKAIENPGLTPSSSLRTALISVFEDAVIDDSSQVH-EELGLEDY 348
NSNK F+DM+DLNFLMCKAIENP LTPSSSLRTA IS+FED VID+S + LG++DY
Sbjct: 357 NSNKQFTDMSDLNFLMCKAIENPNLTPSSSLRTAFISIFEDPVIDESPEPELASLGVQDY 416
Query: 347 VGCASTHGVGPSVAIFDTVRNGKLDCACIYPSPLHSREQIQGLVDDMKRILVEGCNS 177
+GCAS HGVGPSVA+FD +R+GKLDCA +YPSPLHSREQ+ GL+ MK IL+EG S
Sbjct: 417 IGCASIHGVGPSVAVFDALRDGKLDCAFVYPSPLHSREQMDGLIQHMKTILLEGSAS 473
>dbj|BAB63795.1| P0423B08.4 [Oryza sativa (japonica cultivar-group)]
gi|20521257|dbj|BAB91773.1| P0679C12.25 [Oryza sativa
(japonica cultivar-group)]
Length = 490
Score = 143 bits (360), Expect = 1e-33
Identities = 63/121 (52%), Positives = 96/121 (79%)
Frame = -2
Query: 530 AMNSNKHFSDMNDLNFLMCKAIENPGLTPSSSLRTALISVFEDAVIDDSSQVHEELGLED 351
A N+ KH +D+ DLNFLMC+AIENP LT S+LRTA++SVFE+ V+ D S + + G+E+
Sbjct: 364 AKNNKKHLTDIADLNFLMCRAIENPQLTTGSALRTAVVSVFEEPVVYDLSDLQSKAGVEE 423
Query: 350 YVGCASTHGVGPSVAIFDTVRNGKLDCACIYPSPLHSREQIQGLVDDMKRILVEGCNSEN 171
+V CA+ HGVGPS+ +FD++R+G+L+ AC+YP PLHSR+Q+Q +++ +K+IL EG ++
Sbjct: 424 FVCCATVHGVGPSIGLFDSIRDGQLEFACMYPCPLHSRKQMQEILNKVKQILHEGSIGDD 483
Query: 170 Q 168
+
Sbjct: 484 E 484
>ref|NP_494910.1| Trypsin precursor [Caenorhabditis elegans] gi|7511199|pir||T27906
hypothetical protein ZK546.15 - Caenorhabditis elegans
gi|868212|gb|AAA68746.1| Hypothetical protein ZK546.15
[Caenorhabditis elegans]
Length = 331
Score = 34.7 bits (78), Expect = 0.72
Identities = 28/91 (30%), Positives = 42/91 (45%), Gaps = 10/91 (10%)
Frame = +2
Query: 131 SPHALKYTI---NRLIDFHC----YNPPLVFSSYHPLVLESA---PLNEGVMDIYKHNPT 280
SPH+ +T+ +RL C +P V ++ H ES P N + D Y PT
Sbjct: 65 SPHSWPWTVQLLSRLGHHRCGGSLIDPNFVLTAAHCFAKESVNSRPANLFI-DFYSRRPT 123
Query: 281 SHSVQCRIWLRMGQHRVWKHNPHSLPVQALH 373
S+SV R+G HR +PH + ++H
Sbjct: 124 SYSV------RVGGHRSGSGSPHRVTAVSIH 148
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 459,548,034
Number of Sequences: 1393205
Number of extensions: 10123936
Number of successful extensions: 32404
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 30546
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32115
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17596710992
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)