Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
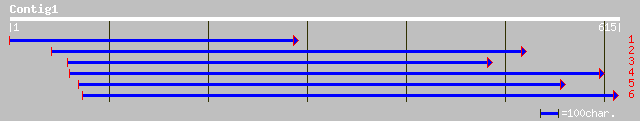
Query= KMC005034A_C01 KMC005034A_c01
(603 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P49199|RS8_ORYSA 40S RIBOSOMAL PROTEIN S8 gi|7440236|pir||T04... 101 8e-21
sp|Q08069|RS8_MAIZE 40S ribosomal protein S8 gi|7440237|pir||T04... 97 1e-19
sp|O81361|RS8_PRUAR 40S ribosomal protein S8 gi|3264759|gb|AAC24... 94 2e-18
ref|NP_197529.1| putative protein; protein id: At5g20290.1, supp... 92 7e-18
ref|NP_200732.1| 40S ribosomal protein S8 - like; protein id: At... 90 3e-17
>sp|P49199|RS8_ORYSA 40S RIBOSOMAL PROTEIN S8 gi|7440236|pir||T04082 probable ribosomal
protein S8 - rice gi|968902|dbj|BAA07207.1| ribosomal
protein S8 [Oryza sativa (japonica cultivar-group)]
Length = 220
Score = 101 bits (251), Expect = 8e-21
Identities = 46/67 (68%), Positives = 55/67 (81%)
Frame = -2
Query: 602 NHVLRKLEKRTENRTLDAHIEEQFGGGRLLACISSRPGQCGRSDGYILEGQRARILHEEA 423
NHV+RKLEKR + RTLDAHIEEQFG GRLLACISSRPGQCGR+DGYILEG+ ++
Sbjct: 151 NHVVRKLEKRQQGRTLDAHIEEQFGSGRLLACISSRPGQCGRADGYILEGKELEFYMKKL 210
Query: 422 SEEEGQG 402
++G+G
Sbjct: 211 QRKKGKG 217
Score = 39.7 bits (91), Expect = 0.030
Identities = 17/21 (80%), Positives = 20/21 (94%)
Frame = -3
Query: 457 KAKELEFYMKKLQKKKGKGAA 395
+ KELEFYMKKLQ+KKGKGA+
Sbjct: 199 EGKELEFYMKKLQRKKGKGAS 219
>sp|Q08069|RS8_MAIZE 40S ribosomal protein S8 gi|7440237|pir||T04088 ribosomal protein
S8 - maize gi|1498053|gb|AAB06330.1| ribosomal protein
S8
Length = 221
Score = 97.4 bits (241), Expect = 1e-19
Identities = 45/66 (68%), Positives = 52/66 (78%)
Frame = -2
Query: 602 NHVLRKLEKRTENRTLDAHIEEQFGGGRLLACISSRPGQCGRSDGYILEGQRARILHEEA 423
NHV RKLEKR E RTLD HIEEQFG GRLLACISSRPGQCGR+DGYILEG+ ++
Sbjct: 152 NHVTRKLEKRKEGRTLDPHIEEQFGSGRLLACISSRPGQCGRADGYILEGKELEFYMKKL 211
Query: 422 SEEEGQ 405
++G+
Sbjct: 212 QRKKGK 217
Score = 37.0 bits (84), Expect = 0.20
Identities = 16/20 (80%), Positives = 18/20 (90%)
Frame = -3
Query: 457 KAKELEFYMKKLQKKKGKGA 398
+ KELEFYMKKLQ+KKGK A
Sbjct: 200 EGKELEFYMKKLQRKKGKSA 219
>sp|O81361|RS8_PRUAR 40S ribosomal protein S8 gi|3264759|gb|AAC24583.1| 40S ribosomal
protein S8 [Prunus armeniaca]
Length = 221
Score = 93.6 bits (231), Expect = 2e-18
Identities = 43/67 (64%), Positives = 53/67 (78%)
Frame = -2
Query: 602 NHVLRKLEKRTENRTLDAHIEEQFGGGRLLACISSRPGQCGRSDGYILEGQRARILHEEA 423
NHV KLEKR + RTLDAHIEEQFGGG+LLACISSRPGQCG++DG ILEG+ ++
Sbjct: 150 NHVAIKLEKRQQGRTLDAHIEEQFGGGKLLACISSRPGQCGKADGDILEGKELEFYMKKL 209
Query: 422 SEEEGQG 402
++G+G
Sbjct: 210 QRKKGKG 216
Score = 39.3 bits (90), Expect = 0.040
Identities = 17/20 (85%), Positives = 19/20 (95%)
Frame = -3
Query: 457 KAKELEFYMKKLQKKKGKGA 398
+ KELEFYMKKLQ+KKGKGA
Sbjct: 198 EGKELEFYMKKLQRKKGKGA 217
>ref|NP_197529.1| putative protein; protein id: At5g20290.1, supported by cDNA:
29997., supported by cDNA: gi_15293006, supported by
cDNA: gi_15450473, supported by cDNA: gi_16974464,
supported by cDNA: gi_20258878 [Arabidopsis thaliana]
gi|15293007|gb|AAK93614.1| unknown protein [Arabidopsis
thaliana] gi|15450474|gb|AAK96530.1| AT5g20290/F5O24_180
[Arabidopsis thaliana] gi|16974465|gb|AAL31236.1|
AT5g20290/F5O24_180 [Arabidopsis thaliana]
gi|20258879|gb|AAM14111.1| unknown protein [Arabidopsis
thaliana] gi|21592577|gb|AAM64526.1| 40S ribosomal
protein S8-like [Arabidopsis thaliana]
Length = 222
Score = 91.7 bits (226), Expect = 7e-18
Identities = 40/67 (59%), Positives = 53/67 (78%)
Frame = -2
Query: 602 NHVLRKLEKRTENRTLDAHIEEQFGGGRLLACISSRPGQCGRSDGYILEGQRARILHEEA 423
NH+LRK+ R E R+LD+HIE+QF GRLLACISSRPGQCGR+DGYILEG+ ++
Sbjct: 154 NHLLRKIASRQEGRSLDSHIEDQFASGRLLACISSRPGQCGRADGYILEGKELEFYMKKI 213
Query: 422 SEEEGQG 402
+++G+G
Sbjct: 214 QKKKGKG 220
Score = 41.2 bits (95), Expect = 0.010
Identities = 18/21 (85%), Positives = 20/21 (94%)
Frame = -3
Query: 457 KAKELEFYMKKLQKKKGKGAA 395
+ KELEFYMKK+QKKKGKGAA
Sbjct: 202 EGKELEFYMKKIQKKKGKGAA 222
>ref|NP_200732.1| 40S ribosomal protein S8 - like; protein id: At5g59240.1
[Arabidopsis thaliana]
Length = 219
Score = 89.7 bits (221), Expect = 3e-17
Identities = 40/66 (60%), Positives = 51/66 (76%)
Frame = -2
Query: 602 NHVLRKLEKRTENRTLDAHIEEQFGGGRLLACISSRPGQCGRSDGYILEGQRARILHEEA 423
NHV RKLE R E R LD+H+EEQF GRLLACI+SRPGQCGR+DGYILEG+ ++
Sbjct: 149 NHVQRKLEMRQEGRALDSHLEEQFSSGRLLACIASRPGQCGRADGYILEGKELEFYMKKL 208
Query: 422 SEEEGQ 405
+++G+
Sbjct: 209 QKKKGK 214
Score = 38.1 bits (87), Expect = 0.088
Identities = 17/20 (85%), Positives = 18/20 (90%)
Frame = -3
Query: 457 KAKELEFYMKKLQKKKGKGA 398
+ KELEFYMKKLQKKKGK A
Sbjct: 197 EGKELEFYMKKLQKKKGKNA 216
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,992,650
Number of Sequences: 1393205
Number of extensions: 10123205
Number of successful extensions: 24427
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 23519
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24391
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23711793746
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)