Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005030A_C03 KMC005030A_c03
(805 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
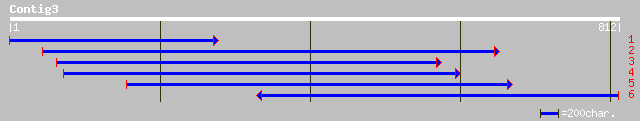
ref|NP_010701.1| Protein required for cell viability; Ydr413cp [... 34 2.2
ref|NP_347317.1| Predicted membrane protein [Clostridium acetobu... 34 2.2
pir||T01578 probable membrane protein DAD1 - maize gi|3264592|gb... 33 3.7
gb|ZP_00037007.1| hypothetical protein [Enterococcus faecium] 33 4.9
ref|ZP_00062647.1| hypothetical protein [Leuconostoc mesenteroid... 33 6.4
>ref|NP_010701.1| Protein required for cell viability; Ydr413cp [Saccharomyces
cerevisiae] gi|2131475|pir||S69735 hypothetical protein
YDR413c - yeast (Saccharomyces cerevisiae)
gi|2194165|gb|AAB64892.1| Ydr413cp [Saccharomyces
cerevisiae]
Length = 191
Score = 34.3 bits (77), Expect = 2.2
Identities = 22/62 (35%), Positives = 30/62 (47%), Gaps = 13/62 (20%)
Frame = -1
Query: 388 STCQPFLPPFFSSTVLAK----SLKAPACSVLNGLRSCRKYLWSSML---------KCFF 248
STC+ + FF+S++ A SL CS + LRSCR + SS+ C F
Sbjct: 129 STCKSSVLSFFASSISASKFKLSLNVFNCSSITSLRSCRIFCLSSIFLSLSCSLINSCAF 188
Query: 247 FC 242
FC
Sbjct: 189 FC 190
>ref|NP_347317.1| Predicted membrane protein [Clostridium acetobutylicum]
gi|25499700|pir||F96983 probable membrane protein
[imported] - Clostridium acetobutylicum
gi|15023557|gb|AAK78657.1|AE007583_4 Predicted membrane
protein [Clostridium acetobutylicum]
Length = 216
Score = 34.3 bits (77), Expect = 2.2
Identities = 17/73 (23%), Positives = 39/73 (53%), Gaps = 2/73 (2%)
Frame = +2
Query: 593 LVLLINIFLLAFSKKMYINSVIAVLAIFLCCVFAALLLLCTSPSVSYFFFV--AYL*SLG 766
+++ IN+F+ +F + I ++ L + + LL++C S++Y +F+ L +
Sbjct: 14 VLMCINLFIDSFKCPVKIRRILRGLVFLIMLRYVTLLIMCLKKSITYIYFMRPIVLLDIL 73
Query: 767 WHPLCLFIISLVY 805
+ PL + I+ V+
Sbjct: 74 YIPLVILIMIFVF 86
>pir||T01578 probable membrane protein DAD1 - maize gi|3264592|gb|AAC24567.1|
FalseDAD1 [Zea mays]
Length = 265
Score = 33.5 bits (75), Expect = 3.7
Identities = 22/67 (32%), Positives = 34/67 (49%)
Frame = +2
Query: 515 NSVAWHSYLHRYHCIDCILYFRGEAPLVLLINIFLLAFSKKMYINSVIAVLAIFLCCVFA 694
N +AW + H YHC C Y L+ IN+ + FSKK NS+ +++ + L +
Sbjct: 48 NLIAWMLWHHSYHCNFCKPYVL--QWLIGYINLLAVIFSKKSK-NSLSSIMVLRLRTSYL 104
Query: 695 ALLLLCT 715
L+L T
Sbjct: 105 CALVLST 111
>gb|ZP_00037007.1| hypothetical protein [Enterococcus faecium]
Length = 299
Score = 33.1 bits (74), Expect = 4.9
Identities = 21/62 (33%), Positives = 35/62 (55%)
Frame = +2
Query: 557 IDCILYFRGEAPLVLLINIFLLAFSKKMYINSVIAVLAIFLCCVFAALLLLCTSPSVSYF 736
I+ ++YF LV I FLL+F++K+Y NS I ++++ +C + + L SV
Sbjct: 99 IEQLIYFVFIFLLVKWIAPFLLSFAEKVYANSSIIIVSLVICLGMSYVADLVGLSSVIGA 158
Query: 737 FF 742
FF
Sbjct: 159 FF 160
>ref|ZP_00062647.1| hypothetical protein [Leuconostoc mesenteroides subsp.
mesenteroides ATCC 8293]
Length = 222
Score = 32.7 bits (73), Expect = 6.4
Identities = 23/61 (37%), Positives = 37/61 (59%), Gaps = 11/61 (18%)
Frame = +2
Query: 593 LVLLINIFLLAFSKKMYINSVIAVLAIFLCCV-------FAALLLLCTSPSV----SYFF 739
LVLLI+I + AF+K +Y+N + V++I + V FA LL++ P+V S++F
Sbjct: 9 LVLLISIEI-AFTKSVYLNLFLIVVSILILIVTKSHLKTFARLLIVSILPAVGSFTSFYF 67
Query: 740 F 742
F
Sbjct: 68 F 68
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 661,934,962
Number of Sequences: 1393205
Number of extensions: 14787007
Number of successful extensions: 40325
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 38359
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40252
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40896270532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)