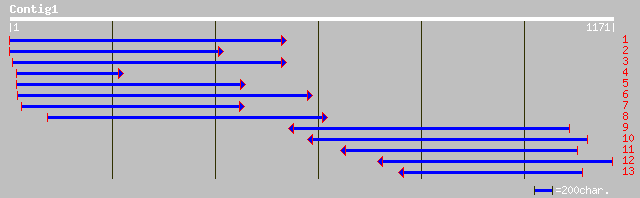
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005022A_C01 KMC005022A_c01
(1165 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_197279.1| NADH-cytochrome b5 reductase; protein id: At5g1... 431 e-124
gb|AAL36459.1|AF205603_1 cytochrome b5 reductase isoform II [Zea... 431 e-123
gb|AAD17694.1| cytochrome b5 reductase [Zea mays] 422 e-121
gb|AAL24304.1| NADH-cytochrome b5 reductase [Arabidopsis thalian... 306 5e-82
dbj|BAA85586.1| NADH-cytochrome b5 reductase [Mortierella alpina... 257 2e-67
>ref|NP_197279.1| NADH-cytochrome b5 reductase; protein id: At5g17770.1, supported by
cDNA: 17432., supported by cDNA: gi_4240115 [Arabidopsis
thaliana] gi|25284885|pir||T52470 cytochrome-b5
reductase (EC 1.6.2.2) [validated] - Arabidopsis
thaliana gi|4240116|dbj|BAA74837.1| NADH-cytochrome b5
reductase [Arabidopsis thaliana]
gi|4240118|dbj|BAA74838.1| NADH-cytochrome b5 reductase
[Arabidopsis thaliana] gi|9759054|dbj|BAB09576.1|
NADH-cytochrome b5 reductase [Arabidopsis thaliana]
gi|21553853|gb|AAM62946.1| NADH-cytochrome b5 reductase
[Arabidopsis thaliana]
Length = 281
Score = 431 bits (1108), Expect(2) = e-124
Identities = 206/240 (85%), Positives = 222/240 (91%)
Frame = -3
Query: 962 LDPDNVKEFKLVKKAQLSHNVAKFTFALPTPTSVLGLPIGQHISGKGKDPQGDEVIKPYT 783
LDP+N KEFKLVK+ QLSHNVAKF F LPT TSVLGLPIGQHIS +GKD QG++VIKPYT
Sbjct: 42 LDPENFKEFKLVKRHQLSHNVAKFVFELPTSTSVLGLPIGQHISCRGKDGQGEDVIKPYT 101
Query: 782 PTTLDSDVGHFELVIKMYPQGRMSHHFREMRVGDYLAVKGPKGRFKYQPNDVRAFGMLAG 603
PTTLDSDVG FELVIKMYPQGRMSHHFREMRVGD+LAVKGPKGRFKYQP RAFGMLAG
Sbjct: 102 PTTLDSDVGRFELVIKMYPQGRMSHHFREMRVGDHLAVKGPKGRFKYQPGQFRAFGMLAG 161
Query: 602 GSGITPMFQVARAILENPNDRTKVHLIYANVTYEDILLKEELDGLATNYPDSFKIYYVLN 423
GSGITPMFQVARAILENP D+TKVHLIYANVTY+DILLKEEL+GL TNYP+ FKI+YVLN
Sbjct: 162 GSGITPMFQVARAILENPTDKTKVHLIYANVTYDDILLKEELEGLTTNYPEQFKIFYVLN 221
Query: 422 QPPEVWDGGVGFVSKEMIEAHLPAPAHDVKILRCGPPPMNKAMAAHLEALEYAPEMQFQF 243
QPPEVWDGGVGFVSKEMI+ H PAPA D++ILRCGPPPMNKAMAA+LEAL Y+PEMQFQF
Sbjct: 222 QPPEVWDGGVGFVSKEMIQTHCPAPASDIQILRCGPPPMNKAMAANLEALGYSPEMQFQF 281
Score = 38.5 bits (88), Expect(2) = e-124
Identities = 22/41 (53%), Positives = 29/41 (70%), Gaps = 1/41 (2%)
Frame = -2
Query: 1074 DFLQIPENQIIVGVAVAIVAVGVG-AYYLYSSKKPKG*LGP 955
+FL+ + QI++GV VA VAVG G AY+L SSKK + L P
Sbjct: 4 EFLRTLDRQILLGVFVAFVAVGAGAAYFLTSSKKRRVCLDP 44
>gb|AAL36459.1|AF205603_1 cytochrome b5 reductase isoform II [Zea mays]
Length = 279
Score = 431 bits (1109), Expect(2) = e-123
Identities = 204/246 (82%), Positives = 221/246 (88%)
Frame = -3
Query: 980 RSPKGNLDPDNVKEFKLVKKAQLSHNVAKFTFALPTPTSVLGLPIGQHISGKGKDPQGDE 801
R P+G LDP+N KEFKLV+K QLSHNVAKF FALPTPTSVLGLPIGQHIS +G+D G+E
Sbjct: 34 RKPRGCLDPENFKEFKLVEKKQLSHNVAKFKFALPTPTSVLGLPIGQHISCRGQDASGEE 93
Query: 800 VIKPYTPTTLDSDVGHFELVIKMYPQGRMSHHFREMRVGDYLAVKGPKGRFKYQPNDVRA 621
VIKPYTPTTLDSD+G FELVIKMYPQGRMSHHFRE +VGDY++VKGPKGRFKY P VRA
Sbjct: 94 VIKPYTPTTLDSDIGSFELVIKMYPQGRMSHHFRETKVGDYMSVKGPKGRFKYLPGQVRA 153
Query: 620 FGMLAGGSGITPMFQVARAILENPNDRTKVHLIYANVTYEDILLKEELDGLATNYPDSFK 441
FGM+AGGSGITPMFQV RAILENP D TKVHLIYANVTYEDILLKEELDG+A NYPD FK
Sbjct: 154 FGMVAGGSGITPMFQVTRAILENPKDNTKVHLIYANVTYEDILLKEELDGMAKNYPDRFK 213
Query: 440 IYYVLNQPPEVWDGGVGFVSKEMIEAHLPAPAHDVKILRCGPPPMNKAMAAHLEALEYAP 261
IYYVLNQPPEVWDGGVGFVSKEMI+ H PAPA D+++LRCGPPPMNKAMAAHL+ L Y
Sbjct: 214 IYYVLNQPPEVWDGGVGFVSKEMIQTHCPAPAADIQVLRCGPPPMNKAMAAHLDGLGYTK 273
Query: 260 EMQFQF 243
EMQFQF
Sbjct: 274 EMQFQF 279
Score = 35.4 bits (80), Expect(2) = e-123
Identities = 19/42 (45%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Frame = -2
Query: 1077 MDFLQIPENQIIVGVAVAIVAVGVGAYYLY-SSKKPKG*LGP 955
M+FLQ + + VA A+VAV G+ YL+ S+KP+G L P
Sbjct: 1 MEFLQGQRLETTLAVAAAVVAVAAGSAYLFLRSRKPRGCLDP 42
>gb|AAD17694.1| cytochrome b5 reductase [Zea mays]
Length = 279
Score = 422 bits (1084), Expect(2) = e-121
Identities = 200/246 (81%), Positives = 221/246 (89%)
Frame = -3
Query: 980 RSPKGNLDPDNVKEFKLVKKAQLSHNVAKFTFALPTPTSVLGLPIGQHISGKGKDPQGDE 801
R PKG LDP+N ++FKLV+K Q+SHNVA+F FALPTPTSVLGLPIGQHIS +G+D G+E
Sbjct: 34 RKPKGCLDPENFRKFKLVEKKQISHNVARFKFALPTPTSVLGLPIGQHISCRGQDATGEE 93
Query: 800 VIKPYTPTTLDSDVGHFELVIKMYPQGRMSHHFREMRVGDYLAVKGPKGRFKYQPNDVRA 621
VIKPYTPTTLDSD+G+FELVIKMYPQGRMSHHFREM+VGDYL+VKGPKGRFKY VRA
Sbjct: 94 VIKPYTPTTLDSDLGYFELVIKMYPQGRMSHHFREMKVGDYLSVKGPKGRFKYHVGQVRA 153
Query: 620 FGMLAGGSGITPMFQVARAILENPNDRTKVHLIYANVTYEDILLKEELDGLATNYPDSFK 441
FGMLAGGSGITPMFQVARAILENPND TKVHLIYANVTYEDILLK+ELD +A YP FK
Sbjct: 154 FGMLAGGSGITPMFQVARAILENPNDNTKVHLIYANVTYEDILLKDELDDMAKTYPGRFK 213
Query: 440 IYYVLNQPPEVWDGGVGFVSKEMIEAHLPAPAHDVKILRCGPPPMNKAMAAHLEALEYAP 261
IYYVLNQPPE W+GGVGFVSKEMI++H PAPA D++ILRCGPPPMNKAMAAHL+ L Y
Sbjct: 214 IYYVLNQPPENWNGGVGFVSKEMIQSHCPAPAEDIQILRCGPPPMNKAMAAHLDELNYTK 273
Query: 260 EMQFQF 243
EMQFQF
Sbjct: 274 EMQFQF 279
Score = 38.1 bits (87), Expect(2) = e-121
Identities = 22/42 (52%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Frame = -2
Query: 1077 MDFLQIPENQIIVGVAVAIVAVGV-GAYYLYSSKKPKG*LGP 955
MDFLQ + V VAVA+ AV GA+ L S+KPKG L P
Sbjct: 1 MDFLQEQSVETTVAVAVAVAAVAAGGAFLLLRSRKPKGCLDP 42
>gb|AAL24304.1| NADH-cytochrome b5 reductase [Arabidopsis thaliana]
gi|18377462|gb|AAL66897.1| NADH-cytochrome b5 reductase
[Arabidopsis thaliana]
Length = 164
Score = 306 bits (783), Expect = 5e-82
Identities = 143/164 (87%), Positives = 154/164 (93%)
Frame = -3
Query: 734 MYPQGRMSHHFREMRVGDYLAVKGPKGRFKYQPNDVRAFGMLAGGSGITPMFQVARAILE 555
MYPQGRMSHHFREMRVGD+LAVKGPKGRFKYQP RAFGMLAGGSGITPMFQVARAILE
Sbjct: 1 MYPQGRMSHHFREMRVGDHLAVKGPKGRFKYQPGQFRAFGMLAGGSGITPMFQVARAILE 60
Query: 554 NPNDRTKVHLIYANVTYEDILLKEELDGLATNYPDSFKIYYVLNQPPEVWDGGVGFVSKE 375
NP D+TKVHLIYANVTY+DILLKEEL+GL TNYP+ FKI+YVLNQPPEVWDGGVGFVSKE
Sbjct: 61 NPTDKTKVHLIYANVTYDDILLKEELEGLTTNYPEQFKIFYVLNQPPEVWDGGVGFVSKE 120
Query: 374 MIEAHLPAPAHDVKILRCGPPPMNKAMAAHLEALEYAPEMQFQF 243
MI+ H PAPA D++ILRCGPPPMNKAMAA+LEAL Y+PEMQFQF
Sbjct: 121 MIQTHCPAPASDIQILRCGPPPMNKAMAANLEALGYSPEMQFQF 164
>dbj|BAA85586.1| NADH-cytochrome b5 reductase [Mortierella alpina]
gi|6070344|dbj|BAA85587.1| NADH-cytochrome b5 reductase
[Mortierella alpina]
Length = 298
Score = 257 bits (657), Expect = 2e-67
Identities = 128/249 (51%), Positives = 169/249 (67%)
Frame = -3
Query: 1013 WGLVLTTSIHLRSPKGNLDPDNVKEFKLVKKAQLSHNVAKFTFALPTPTSVLGLPIGQHI 834
W VL +++P +DP ++FKLV K S N A + FALP +L LPIGQHI
Sbjct: 38 WARVLFKKTAVKTPP--MDPKEYRKFKLVDKVHCSPNTAMYKFALPHEDDLLNLPIGQHI 95
Query: 833 SGKGKDPQGDEVIKPYTPTTLDSDVGHFELVIKMYPQGRMSHHFREMRVGDYLAVKGPKG 654
S + G ++ + YTPT+ DVGHF L IK YPQG +S F E+ +GD + +GPKG
Sbjct: 96 SIMA-NINGKDISRSYTPTSSSDDVGHFVLCIKSYPQGNISKMFSELSIGDSINARGPKG 154
Query: 653 RFKYQPNDVRAFGMLAGGSGITPMFQVARAILENPNDRTKVHLIYANVTYEDILLKEELD 474
+F Y PN RA GM+AGG+G+TPM Q+ RAI++NP D+T+V+ I+ANVT EDI+LK ELD
Sbjct: 155 QFSYTPNMCRAIGMIAGGTGLTPMLQIIRAIVKNPEDKTQVNFIFANVTEEDIILKAELD 214
Query: 473 GLATNYPDSFKIYYVLNQPPEVWDGGVGFVSKEMIEAHLPAPAHDVKILRCGPPPMNKAM 294
L+ +P FK+YYVLN PE W GGVGFV+ +MI+ H+PAPA D+K+L CGPPPM AM
Sbjct: 215 LLSQKHP-QFKVYYVLNNAPEGWTGGVGFVNADMIKEHMPAPAADIKVLLCGPPPMVSAM 273
Query: 293 AAHLEALEY 267
+ + L Y
Sbjct: 274 SKITQDLGY 282
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,083,477,011
Number of Sequences: 1393205
Number of extensions: 26208364
Number of successful extensions: 77379
Number of sequences better than 10.0: 607
Number of HSP's better than 10.0 without gapping: 69927
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 76375
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 71929118964
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)