Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
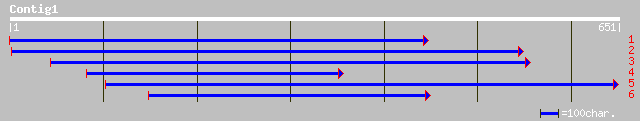
Query= KMC005021A_C01 KMC005021A_c01
(650 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM64471.1| unknown [Arabidopsis thaliana] 110 2e-23
ref|NP_568200.1| putative protein; protein id: At5g08540.1, supp... 110 2e-23
dbj|BAB10003.1| gene_id:MAH20.10~pir||T27191~similar to unknown ... 96 4e-19
ref|NP_196077.1| putative protein; protein id: At5g04570.1 [Arab... 38 0.14
gb|AAM77215.1| DEMETER protein [Arabidopsis thaliana] 38 0.14
>gb|AAM64471.1| unknown [Arabidopsis thaliana]
Length = 346
Score = 110 bits (275), Expect = 2e-23
Identities = 53/89 (59%), Positives = 71/89 (79%), Gaps = 7/89 (7%)
Frame = -1
Query: 650 KGFKRKLAVQTLFGKVFILSELPEFCSRDSSLVVKETFGVTDEDADKLRIHTISDAGNFD 471
KGFKRKLAVQ LFGK++ LSELP+FCS+D+SL+VKE FGVTDEDA+KLRIH +++AG+ +
Sbjct: 255 KGFKRKLAVQALFGKIYYLSELPDFCSKDNSLIVKEIFGVTDEDAEKLRIHALAEAGDIE 314
Query: 470 ALEKMMD-------NSDSEDSSEASFDAS 405
ALEKM++ +SD E+ S D++
Sbjct: 315 ALEKMVEFEKTGESSSDKEEDSNEEDDSN 343
>ref|NP_568200.1| putative protein; protein id: At5g08540.1, supported by cDNA:
249722., supported by cDNA: gi_14334821, supported by
cDNA: gi_15293202 [Arabidopsis thaliana]
gi|14334822|gb|AAK59589.1| unknown protein [Arabidopsis
thaliana] gi|15293203|gb|AAK93712.1| unknown protein
[Arabidopsis thaliana]
Length = 346
Score = 110 bits (275), Expect = 2e-23
Identities = 53/89 (59%), Positives = 71/89 (79%), Gaps = 7/89 (7%)
Frame = -1
Query: 650 KGFKRKLAVQTLFGKVFILSELPEFCSRDSSLVVKETFGVTDEDADKLRIHTISDAGNFD 471
KGFKRKLAVQ LFGK++ LSELP+FCS+D+SL+VKE FGVTDEDA+KLRIH +++AG+ +
Sbjct: 255 KGFKRKLAVQALFGKIYYLSELPDFCSKDNSLIVKEIFGVTDEDAEKLRIHALAEAGDIE 314
Query: 470 ALEKMMD-------NSDSEDSSEASFDAS 405
ALEKM++ +SD E+ S D++
Sbjct: 315 ALEKMVEFEKTAESSSDKEEDSNEEDDSN 343
>dbj|BAB10003.1| gene_id:MAH20.10~pir||T27191~similar to unknown protein
[Arabidopsis thaliana]
Length = 471
Score = 95.9 bits (237), Expect = 4e-19
Identities = 52/112 (46%), Positives = 71/112 (62%), Gaps = 30/112 (26%)
Frame = -1
Query: 650 KGFKRKLAVQTLFGKVFILSELPEFCSRDSSLVVKETFGVTD------------------ 525
KGFKRKLAVQ LFGK++ LSELP+FCS+D+SL+VKE FGVT+
Sbjct: 357 KGFKRKLAVQALFGKIYYLSELPDFCSKDNSLIVKEIFGVTEYVNFILMLLQCSFYLTIS 416
Query: 524 -----EDADKLRIHTISDAGNFDALEKMMD-------NSDSEDSSEASFDAS 405
EDA+KLRIH +++AG+ +ALEKM++ +SD E+ S D++
Sbjct: 417 TKHFSEDAEKLRIHALAEAGDIEALEKMVEFEKTAESSSDKEEDSNEEDDSN 468
>ref|NP_196077.1| putative protein; protein id: At5g04570.1 [Arabidopsis thaliana]
gi|11358330|pir||T48453 hypothetical protein T32M21.170
- Arabidopsis thaliana gi|7406461|emb|CAB85563.1|
putative protein [Arabidopsis thaliana]
Length = 555
Score = 37.7 bits (86), Expect = 0.14
Identities = 28/96 (29%), Positives = 46/96 (47%), Gaps = 1/96 (1%)
Frame = -1
Query: 638 RKLAVQTLFGKVFILSELPEFCSRDSSLVVKETFGVTDEDADKLRIHTISDAGNFDALEK 459
R + V+ G + L+E+P + + E GV ++LR + S F+ LEK
Sbjct: 32 RSVVVEDPEGCILNLNEIPSWQEKVQHPSDMEVSGVDSGSKEQLRDCSNSGIERFNFLEK 91
Query: 458 MMDNSDSED-SSEASFDAS*YKSCSRFLQANRLKGD 354
+ N + E SS+ SFD + ++SC R + K D
Sbjct: 92 SIQNLEEEVLSSQDSFDPAIFQSCGRVGSCSCSKSD 127
>gb|AAM77215.1| DEMETER protein [Arabidopsis thaliana]
Length = 1729
Score = 37.7 bits (86), Expect = 0.14
Identities = 28/96 (29%), Positives = 46/96 (47%), Gaps = 1/96 (1%)
Frame = -1
Query: 638 RKLAVQTLFGKVFILSELPEFCSRDSSLVVKETFGVTDEDADKLRIHTISDAGNFDALEK 459
R + V+ G + L+E+P + + E GV ++LR + S F+ LEK
Sbjct: 810 RSVVVEDPEGCILNLNEIPSWQEKVQHPSDMEVSGVDSGSKEQLRDCSNSGIERFNFLEK 869
Query: 458 MMDNSDSED-SSEASFDAS*YKSCSRFLQANRLKGD 354
+ N + E SS+ SFD + ++SC R + K D
Sbjct: 870 SIQNLEEEVLSSQDSFDPAIFQSCGRVGSCSCSKSD 905
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 520,582,397
Number of Sequences: 1393205
Number of extensions: 10708859
Number of successful extensions: 30337
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 26712
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29640
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27860523586
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)