Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
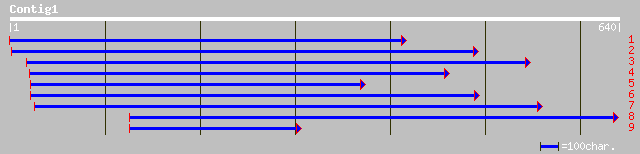
Query= KMC005017A_C01 KMC005017A_c01
(627 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA36972.1| flavonoid 3-O-galactosyl transferase [Vigna mungo] 220 1e-56
dbj|BAA90787.1| UDP glucose: flavonoid 3-O-glucosyltransferase [... 169 2e-41
dbj|BAB41017.1| UDP-glucose:flavonoid 3-O-glucosyltransferase [V... 157 6e-40
dbj|BAB41020.1| UDP-glucose:flavonoid 3-O-glucosyltransferase [V... 156 2e-39
sp|Q40289|UFO7_MANES Flavonol 3-O-glucosyltransferase 7 (UDP-glu... 163 2e-39
>dbj|BAA36972.1| flavonoid 3-O-galactosyl transferase [Vigna mungo]
Length = 455
Score = 220 bits (560), Expect = 1e-56
Identities = 104/150 (69%), Positives = 126/150 (83%)
Frame = -2
Query: 626 SMKDHPKSLLPNGFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVP 447
S+K+H K +LPNGF+ERT GK+V WAPQTQ+LGH SVGVFVTHCGCNSVFES+SNGVP
Sbjct: 308 SLKEHLKGVLPNGFLERTSERGKVVGWAPQTQVLGHGSVGVFVTHCGCNSVFESMSNGVP 367
Query: 446 MICRPFFGDHGMAGRIIEDIWEIGVKIEGGVFTKDGLVKSLNLILVQEEGKIMREKAQKV 267
MICRPFFGDHG+ GR++ED+WEIGV++EGGVFTKDGL+KSL LILV+EEG +M++ A KV
Sbjct: 368 MICRPFFGDHGLTGRMVEDVWEIGVRVEGGVFTKDGLLKSLRLILVEEEGNLMKKNAVKV 427
Query: 266 KKTVLDAARATRESNTGFQDFGGNSLLKCI 177
KKTVLDAA A QDF N+L++ +
Sbjct: 428 KKTVLDAAGA---QGKAAQDF--NTLVELV 452
>dbj|BAA90787.1| UDP glucose: flavonoid 3-O-glucosyltransferase [Ipomoea batatas]
Length = 383
Score = 169 bits (429), Expect = 2e-41
Identities = 78/140 (55%), Positives = 99/140 (70%)
Frame = -2
Query: 626 SMKDHPKSLLPNGFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVP 447
S+K H LP GF+ERTK GKIVPWAPQ Q+L H VGVFVTHCG NS E+IS GV
Sbjct: 235 SLKPHGVKHLPEGFVERTKEFGKIVPWAPQVQVLSHPGVGVFVTHCGWNSTLEAISCGVC 294
Query: 446 MICRPFFGDHGMAGRIIEDIWEIGVKIEGGVFTKDGLVKSLNLILVQEEGKIMREKAQKV 267
MICRPF+GD + R +E +WEIGVKIEGG+FTKDG +K+LN++L + GK+++E K+
Sbjct: 295 MICRPFYGDQKINTRFVESVWEIGVKIEGGIFTKDGTMKALNVVLDSDRGKLLKENVVKL 354
Query: 266 KKTVLDAARATRESNTGFQD 207
K L+A + S FQ+
Sbjct: 355 KGEALEAVKPNGSSTKDFQE 374
>dbj|BAB41017.1| UDP-glucose:flavonoid 3-O-glucosyltransferase [Vitis labrusca x
Vitis vinifera]
Length = 456
Score = 157 bits (398), Expect(2) = 6e-40
Identities = 72/136 (52%), Positives = 102/136 (74%)
Frame = -2
Query: 626 SMKDHPKSLLPNGFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVP 447
S++D + LP GF+E+T+ G +VPWAPQ ++L HE+VG FVTHCG NS++ES++ GVP
Sbjct: 306 SLRDKARVHLPEGFLEKTRGYGMVVPWAPQAEVLAHEAVGAFVTHCGWNSLWESVAGGVP 365
Query: 446 MICRPFFGDHGMAGRIIEDIWEIGVKIEGGVFTKDGLVKSLNLILVQEEGKIMREKAQKV 267
+ICRPFFGD + GR++ED+ EIGV+IEGGVFTK GL+ + IL QE+GK +RE + +
Sbjct: 366 LICRPFFGDQRLNGRMVEDVLEIGVRIEGGVFTKSGLMSCFDQILSQEKGKKLRENLRAL 425
Query: 266 KKTVLDAARATRESNT 219
++T D A + S+T
Sbjct: 426 RETA-DRAVGPKGSST 440
Score = 28.5 bits (62), Expect(2) = 6e-40
Identities = 11/18 (61%), Positives = 16/18 (88%)
Frame = -3
Query: 241 GPQGKATQDFKTLVGIVS 188
GP+G +T++FKTLV +VS
Sbjct: 434 GPKGSSTENFKTLVDLVS 451
>dbj|BAB41020.1| UDP-glucose:flavonoid 3-O-glucosyltransferase [Vitis vinifera]
gi|13620865|dbj|BAB41022.1| UDP-glucose:flavonoid
3-O-glucosyltransferase [Vitis vinifera]
Length = 456
Score = 156 bits (394), Expect(2) = 2e-39
Identities = 72/136 (52%), Positives = 101/136 (73%)
Frame = -2
Query: 626 SMKDHPKSLLPNGFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVP 447
S++D + LP GF+E+T+ G +VPWAPQ ++L HE+VG FVTHCG NS++ES++ GVP
Sbjct: 306 SLRDKARVHLPEGFLEKTRGYGMVVPWAPQAEVLAHEAVGAFVTHCGWNSLWESVAGGVP 365
Query: 446 MICRPFFGDHGMAGRIIEDIWEIGVKIEGGVFTKDGLVKSLNLILVQEEGKIMREKAQKV 267
+ICRPFFGD + GR++ED EIGV+IEGGVFTK GL+ + IL QE+GK +RE + +
Sbjct: 366 LICRPFFGDQRLNGRMVEDALEIGVRIEGGVFTKSGLMSCFDQILSQEKGKKLRENLRAL 425
Query: 266 KKTVLDAARATRESNT 219
++T D A + S+T
Sbjct: 426 RETA-DRAVGPKGSST 440
Score = 28.5 bits (62), Expect(2) = 2e-39
Identities = 11/18 (61%), Positives = 16/18 (88%)
Frame = -3
Query: 241 GPQGKATQDFKTLVGIVS 188
GP+G +T++FKTLV +VS
Sbjct: 434 GPKGSSTENFKTLVDLVS 451
>sp|Q40289|UFO7_MANES Flavonol 3-O-glucosyltransferase 7 (UDP-glucose flavonoid
3-O-glucosyltransferase 7) gi|542017|pir||S41953
UTP-glucose glucosyltransferase - cassava
gi|453253|emb|CAA54614.1| UTP-glucose
glucosyltransferase [Manihot esculenta]
Length = 287
Score = 163 bits (412), Expect = 2e-39
Identities = 74/140 (52%), Positives = 101/140 (71%)
Frame = -2
Query: 626 SMKDHPKSLLPNGFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVP 447
S+KDH K LPNGF++RTK+ G ++ WAPQ +IL H ++GVFVTHCG NS+ ESI GVP
Sbjct: 135 SLKDHSKVHLPNGFLDRTKSHGIVLSWAPQVEILEHAALGVFVTHCGWNSILESIVGGVP 194
Query: 446 MICRPFFGDHGMAGRIIEDIWEIGVKIEGGVFTKDGLVKSLNLILVQEEGKIMREKAQKV 267
MICRPFFGD + GR++ED+WEIG+ ++GGV TK+G + LN IL+Q +GK MRE +++
Sbjct: 195 MICRPFFGDQRLNGRMVEDVWEIGLLMDGGVLTKNGAIDGLNQILLQGKGKKMRENIKRL 254
Query: 266 KKTVLDAARATRESNTGFQD 207
K+ A S+ F +
Sbjct: 255 KELAKGATEPKGSSSKSFTE 274
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 588,714,186
Number of Sequences: 1393205
Number of extensions: 14057815
Number of successful extensions: 36925
Number of sequences better than 10.0: 760
Number of HSP's better than 10.0 without gapping: 35251
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36720
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25586195130
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)