Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
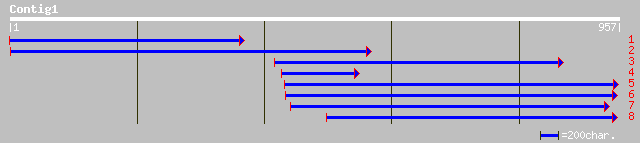
Query= KMC004985A_C01 KMC004985A_c01
(954 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||C96653 hypothetical protein F16P17.2 [imported] - Arabidops... 101 1e-20
ref|NP_176475.1| unknown protein; protein id: At1g62870.1 [Arabi... 100 4e-20
ref|NP_172700.1| hypothetical protein; protein id: At1g12380.1 [... 95 1e-18
pir||D86258 protein F5O11.10 [imported] - Arabidopsis thaliana g... 95 1e-18
gb|AAD48836.1|AF165924_1 auxin-induced basic helix-loop-helix tr... 37 0.45
>pir||C96653 hypothetical protein F16P17.2 [imported] - Arabidopsis thaliana
gi|8493576|gb|AAF75799.1|AC011000_2 Contains weak
similarity to T-type calcium channel isoform from Rattus
norvegicus gb|AF125161. EST gb|R30034 comes from this
gene. [Arabidopsis thaliana]
Length = 762
Score = 101 bits (252), Expect = 1e-20
Identities = 51/78 (65%), Positives = 62/78 (79%), Gaps = 5/78 (6%)
Frame = -3
Query: 952 HATSRGFKCN-----WPLRLGHSRVALERAQKLIFIAAHSKLERRDFSSDEDKDAELFAL 788
HATS GFKCN W G SR A++RAQKLIFI+A+SK ERRDFS++E++DAEL A+
Sbjct: 685 HATSCGFKCNSSVLRWVNSNGRSRAAVDRAQKLIFISANSKFERRDFSNEEERDAELLAM 744
Query: 787 VNGEDDVLNEVFLDTSSV 734
NGEDDVLN+V +DTSSV
Sbjct: 745 ANGEDDVLNDVLIDTSSV 762
>ref|NP_176475.1| unknown protein; protein id: At1g62870.1 [Arabidopsis thaliana]
Length = 796
Score = 100 bits (248), Expect = 4e-20
Identities = 50/77 (64%), Positives = 61/77 (78%), Gaps = 5/77 (6%)
Frame = -3
Query: 952 HATSRGFKCN-----WPLRLGHSRVALERAQKLIFIAAHSKLERRDFSSDEDKDAELFAL 788
HATS GFKCN W G SR A++RAQKLIFI+A+SK ERRDFS++E++DAEL A+
Sbjct: 685 HATSCGFKCNSSVLRWVNSNGRSRAAVDRAQKLIFISANSKFERRDFSNEEERDAELLAM 744
Query: 787 VNGEDDVLNEVFLDTSS 737
NGEDDVLN+V +DTSS
Sbjct: 745 ANGEDDVLNDVLIDTSS 761
>ref|NP_172700.1| hypothetical protein; protein id: At1g12380.1 [Arabidopsis
thaliana]
Length = 793
Score = 95.1 bits (235), Expect = 1e-18
Identities = 47/78 (60%), Positives = 60/78 (76%), Gaps = 5/78 (6%)
Frame = -3
Query: 952 HATSRGFKCN-----WPLRLGHSRVALERAQKLIFIAAHSKLERRDFSSDEDKDAELFAL 788
HAT+ GFKCN W G S A++RAQKLIFI+A+SK ERRDFS++ED+DAEL A+
Sbjct: 716 HATTGGFKCNSSLLKWVNSNGRSHAAVDRAQKLIFISANSKFERRDFSNEEDRDAELLAM 775
Query: 787 VNGEDDVLNEVFLDTSSV 734
NG+D +LN+V +DTSSV
Sbjct: 776 ANGDDHMLNDVLVDTSSV 793
>pir||D86258 protein F5O11.10 [imported] - Arabidopsis thaliana
gi|8778648|gb|AAF79656.1|AC025416_30 F5O11.10
[Arabidopsis thaliana]
Length = 856
Score = 95.1 bits (235), Expect = 1e-18
Identities = 47/78 (60%), Positives = 60/78 (76%), Gaps = 5/78 (6%)
Frame = -3
Query: 952 HATSRGFKCN-----WPLRLGHSRVALERAQKLIFIAAHSKLERRDFSSDEDKDAELFAL 788
HAT+ GFKCN W G S A++RAQKLIFI+A+SK ERRDFS++ED+DAEL A+
Sbjct: 779 HATTGGFKCNSSLLKWVNSNGRSHAAVDRAQKLIFISANSKFERRDFSNEEDRDAELLAM 838
Query: 787 VNGEDDVLNEVFLDTSSV 734
NG+D +LN+V +DTSSV
Sbjct: 839 ANGDDHMLNDVLVDTSSV 856
>gb|AAD48836.1|AF165924_1 auxin-induced basic helix-loop-helix transcription factor
[Gossypium hirsutum]
Length = 314
Score = 37.0 bits (84), Expect = 0.45
Identities = 22/60 (36%), Positives = 27/60 (44%), Gaps = 8/60 (13%)
Frame = +1
Query: 760 HSTHHPHH*PRQTALHPYLHHWKNPSFPI--------SNELR*ISTSAPSPVLPWSVLTA 915
H HH HH Q LH + HH P+FP N+ S+S P P LP S +A
Sbjct: 10 HHHHHSHHHHHQQPLHHHHHHHHRPTFPFQLLEKKEEDNQPCSSSSSLPFPSLPVSSSSA 69
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 778,240,697
Number of Sequences: 1393205
Number of extensions: 16740365
Number of successful extensions: 41554
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 36194
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39729
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 53801056208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)