Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004978A_C01 KMC004978A_c01
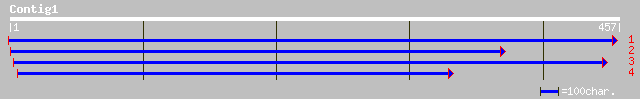
(457 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL24274.1| At2g46910/F14M4.26 [Arabidopsis thaliana] 107 4e-23
ref|NP_182214.2| unknown protein; protein id: At2g46915.1, suppo... 107 4e-23
dbj|BAC22227.1| hypothetical protein~similar to Arabidopsis thal... 97 6e-20
pir||T02196 hypothetical protein At2g46910 [imported] - Arabidop... 49 2e-05
ref|NP_002205.1| integrin, beta 8 [Homo sapiens] gi|124975|sp|P2... 31 5.2
>gb|AAL24274.1| At2g46910/F14M4.26 [Arabidopsis thaliana]
Length = 731
Score = 107 bits (268), Expect = 4e-23
Identities = 48/85 (56%), Positives = 67/85 (78%)
Frame = -3
Query: 455 ILAYAVLLKAEKDQVTSRQSVGEKCERFMYEVFKVKVEMPIDKALNTLLRLSLATETCID 276
IL YA++L+A K+Q S + VG++CERFMY+ FK+KVEM ++KA++TL+RL L TET +D
Sbjct: 646 ILTYAIILQAGKNQNMSYKGVGDRCERFMYDTFKIKVEMRVEKAISTLVRLGLVTETLVD 705
Query: 275 GRHGFLAIPCPQAYVALKERWNSLL 201
A+PCPQAY++LKE W+SLL
Sbjct: 706 SNTKLQAVPCPQAYISLKELWSSLL 730
>ref|NP_182214.2| unknown protein; protein id: At2g46915.1, supported by cDNA:
gi_16604536 [Arabidopsis thaliana]
Length = 495
Score = 107 bits (268), Expect = 4e-23
Identities = 48/85 (56%), Positives = 67/85 (78%)
Frame = -3
Query: 455 ILAYAVLLKAEKDQVTSRQSVGEKCERFMYEVFKVKVEMPIDKALNTLLRLSLATETCID 276
IL YA++L+A K+Q S + VG++CERFMY+ FK+KVEM ++KA++TL+RL L TET +D
Sbjct: 410 ILTYAIILQAGKNQNMSYKGVGDRCERFMYDTFKIKVEMRVEKAISTLVRLGLVTETLVD 469
Query: 275 GRHGFLAIPCPQAYVALKERWNSLL 201
A+PCPQAY++LKE W+SLL
Sbjct: 470 SNTKLQAVPCPQAYISLKELWSSLL 494
>dbj|BAC22227.1| hypothetical protein~similar to Arabidopsis thaliana chromosome 2,
At2g46910/F14M4.26 [Oryza sativa (japonica
cultivar-group)]
Length = 823
Score = 97.4 bits (241), Expect = 6e-20
Identities = 48/85 (56%), Positives = 60/85 (70%)
Frame = -3
Query: 455 ILAYAVLLKAEKDQVTSRQSVGEKCERFMYEVFKVKVEMPIDKALNTLLRLSLATETCID 276
+LAY +LL +K QV+SR S+ + CE+FMYE FK K+EMPIDKA+ TLLRL L E D
Sbjct: 727 LLAYGMLLCRKKYQVSSRVSIRDTCEQFMYEKFKAKIEMPIDKAMETLLRLGLVIELPTD 786
Query: 275 GRHGFLAIPCPQAYVALKERWNSLL 201
G +A+PC AY LK RW+SLL
Sbjct: 787 GGSRVIALPCSDAYEILKSRWDSLL 811
>pir||T02196 hypothetical protein At2g46910 [imported] - Arabidopsis thaliana
gi|20197137|gb|AAM14933.1| unknown protein [Arabidopsis
thaliana]
Length = 734
Score = 48.9 bits (115), Expect = 2e-05
Identities = 20/38 (52%), Positives = 31/38 (80%)
Frame = -3
Query: 425 EKDQVTSRQSVGEKCERFMYEVFKVKVEMPIDKALNTL 312
++ Q S + VG++CERFMY+ FK+KVEM ++KA++TL
Sbjct: 510 DRYQNMSYKGVGDRCERFMYDTFKIKVEMRVEKAISTL 547
>ref|NP_002205.1| integrin, beta 8 [Homo sapiens] gi|124975|sp|P26012|ITB8_HUMAN
Integrin beta-8 precursor gi|87965|pir||A41029 integrin
beta-8 chain precursor - human gi|184521|gb|AAA36034.1|
integrin beta-8 subunit
Length = 769
Score = 31.2 bits (69), Expect = 5.2
Identities = 16/46 (34%), Positives = 21/46 (44%), Gaps = 1/46 (2%)
Frame = -3
Query: 284 CIDGRH-GFLAIPCPQAYVALKERWNSLLC*NPICTFPSVLETTST 150
C D R G CP Y A KE WN + C +P ++L+ T
Sbjct: 614 CTDPRSIGRFCEHCPTCYTACKENWNCMQCLHPHNLSQAILDQCKT 659
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 380,290,989
Number of Sequences: 1393205
Number of extensions: 7751708
Number of successful extensions: 16395
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 16095
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 16394
length of database: 448,689,247
effective HSP length: 112
effective length of database: 292,650,287
effective search space used: 11413361193
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)