Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004969A_C01 KMC004969A_c01
(588 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
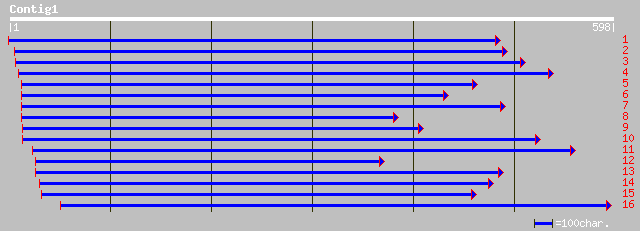
dbj|BAB92170.1| OSJNBa0016I09.22 [Oryza sativa (japonica cultiva... 91 1e-17
ref|NP_174212.1| hypothetical protein; protein id: At1g29170.1 [... 79 4e-14
pir||B86414 hypothetical protein F28N24.14 - Arabidopsis thalian... 79 6e-14
ref|NP_195793.1| putative protein; protein id: At5g01730.1 [Arab... 77 2e-13
gb|AAO22743.1| unknown protein [Arabidopsis thaliana] 73 2e-12
>dbj|BAB92170.1| OSJNBa0016I09.22 [Oryza sativa (japonica cultivar-group)]
Length = 147
Score = 90.9 bits (224), Expect = 1e-17
Identities = 54/116 (46%), Positives = 76/116 (64%), Gaps = 1/116 (0%)
Frame = -3
Query: 532 RRIAKVEQIQSNHDGHESYENMIHQLKEKLDQPKLNGWKEANQLRMG-KETDEREDFLHQ 356
R++ V++I SN E +++ Q+++K Q KLNG +++ + K DERE+ L Q
Sbjct: 33 RKVDGVKEI-SNPLDIEIRASLLQQIRDKSGQQKLNGHEKSKAVGNDTKNLDEREELLQQ 91
Query: 355 IRAKSFNLRPTVTGKPNATTVPTASVKVTAILEKANAIRQVVGSDDGEEDDDTWSD 188
IR+K+FNLR T K N ++ TA+ V AILEKANAIRQ V SD+G DDD+WSD
Sbjct: 92 IRSKTFNLRRTNASKTNTSSPTTANSSVVAILEKANAIRQAVASDEG-GDDDSWSD 146
>ref|NP_174212.1| hypothetical protein; protein id: At1g29170.1 [Arabidopsis thaliana]
Length = 1016
Score = 79.0 bits (193), Expect = 4e-14
Identities = 48/101 (47%), Positives = 61/101 (59%), Gaps = 4/101 (3%)
Frame = -3
Query: 475 ENMIHQLKEKLDQPKL-NGWKEANQLRMGKETDEREDFLHQIRAKSFNLRP---TVTGKP 308
+N + + +P++ N +E Q KET E DFL QIR + FNLRP T T
Sbjct: 918 KNELPSMVTSAPKPEIKNNVREEKQSANAKET-ETGDFLQQIRTQQFNLRPVVMTTTSSA 976
Query: 307 NATTVPTASVKVTAILEKANAIRQVVGSDDGEEDDDTWSDT 185
ATT P + K++AILEKAN+IRQ V S DG+E DTWSDT
Sbjct: 977 TATTDPIINTKISAILEKANSIRQAVASKDGDE-SDTWSDT 1016
>pir||B86414 hypothetical protein F28N24.14 - Arabidopsis thaliana
gi|9502422|gb|AAF88121.1|AC021043_14 Hypothetical protein
[Arabidopsis thaliana]
Length = 1020
Score = 78.6 bits (192), Expect = 6e-14
Identities = 46/84 (54%), Positives = 53/84 (62%), Gaps = 3/84 (3%)
Frame = -3
Query: 427 NGWKEANQLRMGKETDEREDFLHQIRAKSFNLRP---TVTGKPNATTVPTASVKVTAILE 257
N +E Q KET E DFL QIR + FNLRP T T ATT P + K++AILE
Sbjct: 939 NNVREEKQSANAKET-ETGDFLQQIRTQQFNLRPVVMTTTSSATATTDPIINTKISAILE 997
Query: 256 KANAIRQVVGSDDGEEDDDTWSDT 185
KAN+IRQ V S DG+E DTWSDT
Sbjct: 998 KANSIRQAVASKDGDE-SDTWSDT 1020
>ref|NP_195793.1| putative protein; protein id: At5g01730.1 [Arabidopsis thaliana]
gi|11357831|pir||T48194 hypothetical protein F7A7.250 -
Arabidopsis thaliana gi|7327832|emb|CAB82289.1| putative
protein [Arabidopsis thaliana]
Length = 1200
Score = 76.6 bits (187), Expect = 2e-13
Identities = 39/88 (44%), Positives = 53/88 (59%), Gaps = 2/88 (2%)
Frame = -3
Query: 445 LDQPKLNGWKEANQLRMGKETDEREDFLHQIRAKSFNLRPT-VTGKPN-ATTVPTASVKV 272
+D+ L E N+ +G DE + L IR+KSFNLRP +G+PN VP ++KV
Sbjct: 1113 IDRSMLRKVSEGNRTHVGARVDENDSLLEIIRSKSFNLRPADASGRPNFQVAVPKTNLKV 1172
Query: 271 TAILEKANAIRQVVGSDDGEEDDDTWSD 188
AILEKAN +RQ + D E D D+WS+
Sbjct: 1173 AAILEKANTLRQAMAGSDDEHDSDSWSE 1200
>gb|AAO22743.1| unknown protein [Arabidopsis thaliana]
Length = 700
Score = 73.2 bits (178), Expect = 2e-12
Identities = 45/113 (39%), Positives = 60/113 (52%), Gaps = 13/113 (11%)
Frame = -3
Query: 484 ESYENMIHQLKEK------LDQPKL-------NGWKEANQLRMGKETDEREDFLHQIRAK 344
E N + +K+K + PKL N ++ Q E DFLHQIR K
Sbjct: 587 EKNNNSLTAVKKKEPHIVTVSDPKLVTKVHLKNNVRDYKQSHGNTNETEAGDFLHQIRTK 646
Query: 343 SFNLRPTVTGKPNATTVPTASVKVTAILEKANAIRQVVGSDDGEEDDDTWSDT 185
FNLR V K +++ T + ++ ILEKAN+IRQ V SDDGE + DTWSD+
Sbjct: 647 QFNLRRVVRTKTSSSET-TMNTNISVILEKANSIRQAVASDDGEGESDTWSDS 698
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,748,960
Number of Sequences: 1393205
Number of extensions: 10837873
Number of successful extensions: 36438
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 33725
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36292
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)